20.109(S20):Enrich candidate clones using fluorescence-activated cell sorter (FACS) (Day1)
Contents
Introduction
Flow cytometry is a cell analysis technique that has revolutionized biology by allowing researchers to analyze and isolate cells based on their physical and fluorescent qualities. Genetic reporters, labeled antibodies and small molecule tags or dyes may be used to introduce fluorescent signal into cells. You can isolate a pure sample of a particular cell type from a complex mixture of cells by using a modified flow cytometer equipped to perform FACS: Fluorescence Activated Cell Sorting. If the machine is only able to count, and not sort subpopulations of cells, then the procedure is called flow cytometry, while cell sorting is called FACS.
Today we will be using FACS to sort a library of yeast, a library is a collection of DNA fragments, usually on plasmids, that is stored and propagated in a population of microorganisms. A single microorganism carting one plasmid in the library is called a clone. These yeast are expressing a single-chain antibody fragment (scFv) that binds to the enzyme lysozyme. Lysozyme is an abundant, inexpensive and commercially valuable enzyme for the food and pharmaceutical industry. Our goal is to find a clone of a single-chain antibody fragment that has improved binding to lysozyme. We will use fluorescently labeled antibodies to mark yeast that express the scFv and fluorescently labeled streptavidin to mark the presence of bound lysozyme. Using these two fluorescent markers together we will isolate a population of yeast from the library that bind to lysozyme with high affinity.
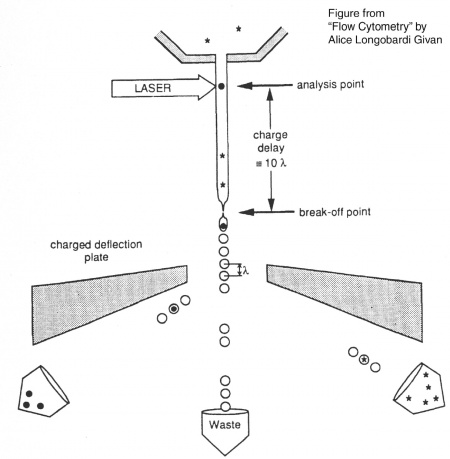
Before researchers had flow cytometers, there were Coulter counters. Coulter counters are automated cell counting machines developed in the 1950s that count cells as they flow in a liquid stream. In an ingenious conceptual leap, Mack Fulwyler combined the technology of ink jet printers with that of Coulter counters to develop the first flow cytometer. The ink jet printer head works by vibrating a nozzle so that a spray of discrete droplets is formed. Similarly, in a flow cytometer, a liquid suspension of cells is forced at high pressure through a vibrating nozzle to create tiny charged droplets, each containing a single cell. The stream of droplets passes in front of a laser beam, and the scattered light is analyzed by a series of filters and photomultiplier tubes that convert the light signal into electrical impulses. Thus, each cell is "interrogated." In FACS, the spectral qualities of the cell are analyzed nearly instantaneously and compared to user-specified spectral qualities. For example, if you have a mixture of green fluorescent cells and non-fluorescent cells, you can ask the machine to isolate the green cells. If a cell registers as green, an electrical charge deflects the cell causing it to fall into a collection chamber. In flow cytometry, each cell is interrogated, and documented as fluorescent or not, but then all the cells go into the same waste stream.Cell sorting, or FACS, is technically challenging and most FACS machines are only run by experts. Although this analysis will primarily be done by the flow core technician, we will briefly describe the main steps to cell sorting. The key to collecting the population of cells you would like is running the appropriate controls to identify these cells accurately on the machine. The first control is a mock/negative control; for our experiment this is unstained yeast. These cells are first analyzed using side scatter versus forward scatter after the cell passes through the analysis point of the laser. Forward scatter (FSC) is a measurement of the sample's refraction of light and therefore often is correlated with cell size. Side scatter (SSC) tells us something about internal complexity of the cell, like size of nucleus or other organelles. The cells should be the largest objects in the solution, while dust and cell debris will tend to fall at very low forward scatter axis. Next we look at our single-color fluorescence staining controls. This allows the user to establish the signal from that single fluorophore and determine if there is overlap in the single-color channels and compensate for that overlap. These samples will be yeast stained with antibodies that include the fluorophore AF488 alone and yeasted stained with streptavidin conjugated to AF647 alone. Finally, we analyze the experimental samples, which are co-stained with AF488(green) labeled antibodies to mark yeast that express the scFv and AF647(far red) labeled streptavidin to mark the presence of bound lysozyme. By collecting the cells that have high green and high far red signal we hope to identify single scFv clones that have improved lysozyme binding.
Flow cytometers have been around since the 1970s, and while the details have gotten fancier (more colors, faster measurements, etc), the general principles have remained the same. It is truly a testament to the versatility and utility of flow cytometry that the technology does not look to be supplanted by another approach any time soon.
Protocols:
The library of yeast you will be sorting today were grown from a population that was tenfold the estimated starting library diversity to assure we will have adequate coverage of the single-chain antibody fragment clones represented in this population. The single-chain antibody fragment expression was induced by growing the yeast in media containing galactose for 20 hours at 20°C.
Part 1: Prepare cells for cell sorting
- Obtain an aliquot of induced yeast library from the 4°C cooler and vortex to resuspend the cells.
- Measure the optical density of the yeast at 600nm using the spectrophotometer.
- Remember to bring media alone as a blank.
- An optical density of 1 at 600nm equals 1x107yeast per mL.
- Estimate the volume needed to have 1x107yeast. In a fresh tube resuspend 1x107 yeast in a final volume of 1mL of PBSA.
- Pellet the yeast at 7,500g for 30sec and remove the supernatant.
- Resuspend the yeast pellet in 1mL fresh PBSA. Pellet the yeast once more and remove the supernatant.
- Resuspend the yeast in 1mL fresh PBSA with 100nM of biotinylated lysozyme.
- The stock of biotinylated lysozyme is 1.87mg/mL and the molecular weight is 14,300. Calculate the volume of stock to dilute into 1mL of PBSA to make a final concentration of 100nM.
- Incubate the yeast library and the biotinylated lysozyme together for 60min at room temperature on the nutator.
- After 60min add 1:1000 anti-c-myc primary antibody to the lysozyme/cell mix, invert 3 times and place the mix back on nutator for an additional 60min at room temp.
- Pellet the cells at 7,500g for 30sec and wash the cells with 1mL ice-cold PBSA.
- Resuspend the cells in 500uL of ice-cold PBSA with 1:1000 AF488 goat anti-chicken IgG secondary antibody and 1:1000 AF647 conjugated to Streptavidin. Leave solution protected from light and on ice for 30min.
- Pellet the cells and resuspend in 1mL of ice-cold PBSA. Leave cells on ice.
Part 2: Fluorescence Activated Cell Sorting
The remainder of the sorting experiment will be carried out by an experienced technician at the Koch Flow Cytometry Core. We'll prepare and hand over the samples as the analysis and sorting is happening. The unstained and single stain control samples were prepared by the teaching faculty and will be analyzed prior to your stained samples. All samples will be carried to the Koch flow core on ice and in secondary containment along with necessary tubes and reagents.
- Place autoclaved or sterile tubes into the FACS collection rack with 1mL of PBSA in each tube.
- After the controls have established which cell population will be collected, quickly vortex the stained yeast and forcefully pipette the cell through the 35µm nylon mesh cap of a Falcon flow cytometry tube.
- This reduces the likelihood of cell aggregates, which can cause critical errors and clogs in the machine.
- Immediately place the cells on the sample injection port (SIP.)
- Cells will begin to flow through the machine and be sorted into the collection tubes as they pass through analyzer and are charged.
- Once the sorting is complete, rinse the tube with 1ml SDCAA and put on ice.
- Transfer the cells back to lab and add additional SDCAA media to bring the volume up to 5mL.
- Leave the cells at 30°C for 2-3 days.
Reagents
- anti-c-myc, chicken IgY fraction (ThermoFisher)
- Alexa Fluor 488 goat anti-chicken IgG (ThermoFisher)
- Alexa Fluor 647 conjugate streptavidin (ThermoFisher)
- PBSA, PBS with 0.1% Albumin (Sigma)
- Falcon flow cytometry filter cap tube (Fisher Scientific)
Next day: Harvest candidate clones and prepare for sequencing