Difference between revisions of "ThorLabs OTKB optical trapping kit"
(→Analysis) |
(→Matlab OTKB GUI) |
||
| (35 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
===Suggested modifications to OTKB layout=== | ===Suggested modifications to OTKB layout=== | ||
| − | |||
| − | + | ''Note: numbers in parenthesis refer to the block diagram on page 4 of the Thorlabs "Optical Trap Application Setup" manual" or Thorlabs part numbers.'' | |
| − | # | + | #The FiberPort Collimator (2) is connected to the Beam Steering Mirror (KCB1) with 4 cage rods (ER05), instead of a lens tube. (Use a 30mm optical cage plate gasket CPG3 to fill the small gap.)The FiberPort Collimator is no longer mounted on a cage plate mounting base (CPB1). |
| − | # | + | #The Beam Steering Mirror is connected to a 0.5" thick 30mm cage plate (CP02T) with 4 ER05 cage rods. Use a 0.3" SM1 lens tube (SM1L03) and an externally threaded tube coupler (SM1T2) to enclose the optical path. |
| − | #Use | + | #Use three 8" cage rods (ER8) rods to connect the CP02T to the 60mm cage cube (C6W) containing the Dichroic Mirror (4). Mount two 30mm cage plates (CP02) on the rods. The one closest to the KCB1 holds a 0.5' SM1 lens tube(SM1L05) that holds first achromatic doublet (AC254-060-B) and the second connected to a lens tube on either side and mounted on a base for stability. Plastic tubing encloses the optical path. |
| − | + | #A SM1L03 lens tube connected to a 1" SM1 lens tube (SM1L10) was screwed into the vertical segment's KCB1. The SM1L05, that holds the second achromatic doublet (part number AC-254-150-B), attached to the C6W, was connected to these combined lens tubes via a 1 inch piece of plastic tubing. | |
| − | + | #The beam path in the vertical segment below the objective was enclosed by adding a SM1L05 lens tube connected to a SM1L03 lens tube threaded into it to the top of the KCB1. In addition, the cage translator (SM1Z) that originally held the objective was replaced by a CP02. This CP02 was mounted on the 4 3" cage rods (ER3) that were attached to the KCB1. | |
| − | # | + | #The sample holder was changed from part AMA-SLH to a custom build slide holder attached to a mounting plate (RB13P1). We also reoriented the direction of the translating breadboard (TBB0606) so that it slides out parallel to the QPD segment. |
| − | # | + | |
| − | # | + | |
#The height of the stage was lowered. | #The height of the stage was lowered. | ||
| − | #Mount the | + | #Mount the(TBB0606) to the breadboard with four 2" long, 1.5" diameter mounting posts (P2). We eliminated the spacers (PS3) and bases (PB2) from each of the mounting legs. |
===Suggested modifications not yet implemented=== | ===Suggested modifications not yet implemented=== | ||
| Line 28: | Line 25: | ||
===Alignment procedure=== | ===Alignment procedure=== | ||
| − | + | ''Note: throughout this procedure, the Relay Lens (3A) will be called the Beam Expander Lens and Relay Lens II (3B) will be called the Tube lens. | |
| − | + | ||
| − | + | #Align and collimate the FiberPort as per section 4.1 of the ThorLabs Optical Trap Application Setup manual. | |
| − | + | #Construct the beam expander segment and vertical segment before proceeding with the alignment. | |
| − | # | + | ##Mount the Beam Expander Lens (3A) and the Tube Lens (4A) in lens tubes, but set these aside instead of mounting the tubes in the cage plates. The position of the Beam Expander Lens will be adjusted by sliding the cage plate it is mounted to. |
| − | # | + | ##Use 3 cage rods instead of 4 in the beam expander section so that lenses can be inserter and removed during the alignment procedure. |
| − | # | + | #Align the Beam Steering Mirror (4) |
| − | + | ##Use two alignment jigs of your choosing ( | |
| + | Follow steps 9-12 in section 4.1 of the ThorLabs OTKB manual except don't attach the ring iris to the KCB1. Instead: | ||
| − | + | #Attach 3 ER8 cage rods to the KCB1. | |
| − | + | ||
| − | + | ||
#Slide 2 CP02s onto the cage rods, positioning one halfway to the end of the rods and the other at the end of the rods. Tighten the CP02s so they can't slide around. | #Slide 2 CP02s onto the cage rods, positioning one halfway to the end of the rods and the other at the end of the rods. Tighten the CP02s so they can't slide around. | ||
#Attach an SM1D12D ring iris to each CP02 and open them all the way. | #Attach an SM1D12D ring iris to each CP02 and open them all the way. | ||
| − | + | #Check your laser collimation and adjust the plunge screws accordingly to re-collimate your beam. | |
| − | + | #Use the alignment techniques stated in the previous section to align the beam | |
| − | + | #Remove the parts attached in steps 1-3. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | #Check your laser collimation | + | |
| − | # | + | |
| − | + | ||
| − | + | ||
| − | #Remove the parts attached in | + | |
====Aligning the First Dichroic and Second KCB1==== | ====Aligning the First Dichroic and Second KCB1==== | ||
| − | + | Complete up to step 10 of section 4.3 in the Thorlabs OTKB manual, skipping the alignment steps in section 4.2 (steps 21-22 and 30-31). It is useful to use 4 ER16 (2 ER8 rods connected together) rods instead of the 4 ER8 rods mentioned in step 11 of section 4.3. Then: | |
| − | + | #Check the collimation of your beam by adjusting the CP02 holding the first achromatic doublet (f = 60mm). Secure this CP02 when you are confident in the laser collimation | |
| − | + | #Close the iris from step 10 in section 4.3 of the OTKB manual. Adjust the rotate the dichroic and adjust its tilt screws so that the beam shoots through the center of the closed iris. | |
| − | #Check the collimation of your beam by adjusting the CP02 holding the first achromatic doublet (f = 60mm) | + | #Open that iris and close the iris mentioned in step 13 in section 4.3 of the OTKB manual. Using the 2 tilt screws on the KCB1, adjust the beam so that it shoots through the center of the closed iris. |
| − | # | + | #Repeat steps 2-3 until the beam goes through the center of both irises when they are closed. |
| − | + | #Remove the parts attached in steps 10-13 in section 4.3 of the OTKB manual | |
| − | + | ||
| − | #Open that iris and close the iris | + | |
| − | #Repeat steps | + | |
| − | + | ||
====Collimating the Objective and Condenser==== | ====Collimating the Objective and Condenser==== | ||
| − | |||
#Install the objective and the rest of the vertical segment as mentioned in section 4.3 of the OTKB manual accounting for the changes in the "Modifications" section. | #Install the objective and the rest of the vertical segment as mentioned in section 4.3 of the OTKB manual accounting for the changes in the "Modifications" section. | ||
#Mount the condenser as described in section 4.4 step 1-3 of the OTKB manual, disregarding the recommended height. | #Mount the condenser as described in section 4.4 step 1-3 of the OTKB manual, disregarding the recommended height. | ||
#Tighten the C1500 clamp holding the condenser such that it doesn't freely slide along the DP14 but it can still be moved very small amounts either by hand or by being tapped by a blunt instrument such as the handle of a screwdriver. | #Tighten the C1500 clamp holding the condenser such that it doesn't freely slide along the DP14 but it can still be moved very small amounts either by hand or by being tapped by a blunt instrument such as the handle of a screwdriver. | ||
| − | #Turn the laser on, load a sample slide (prepared as instructed in section 3.5 of the [http://scripts.mit.edu/~20.309/wiki/images/2/27/Optical_Trapping_Lab_Manual.pdf] using immersion oil, and adjust the condenser height such that the beam is collimated | + | #Turn the laser on, load a sample slide (prepared as instructed in section 3.5 of the [http://scripts.mit.edu/~20.309/wiki/images/2/27/Optical_Trapping_Lab_Manual.pdf] using immersion oil, and adjust the condenser height by tapping on the C1500 clamp that holds it such that the beam is collimated. The condenser will be very close to the objective when properly collimated(on the order of a few mm). |
#Tighten the C1500 clamp holding the condenser completely. | #Tighten the C1500 clamp holding the condenser completely. | ||
====Aligning the Second Dichroic==== | ====Aligning the Second Dichroic==== | ||
| − | |||
#Complete section 4.4 of the OTKB manual (incorporating all necessary changes from the "Modifications" section) | #Complete section 4.4 of the OTKB manual (incorporating all necessary changes from the "Modifications" section) | ||
#Attach the assembly mentioned in step 5 from the "Aligning the First Dichroic and Second KCB1" section to the face of the C6W to which the QPD will eventually be attached. | #Attach the assembly mentioned in step 5 from the "Aligning the First Dichroic and Second KCB1" section to the face of the C6W to which the QPD will eventually be attached. | ||
| Line 88: | Line 66: | ||
====QPD==== | ====QPD==== | ||
Assuming you have accurately aligned your trap, the laser beam should shine on a quadrant of the QPD. Using the two screws on the HPT1 cage mount, adjust the beam such that it shines on the center of the QPD. Congratulations, you have now aligned your optical trap! | Assuming you have accurately aligned your trap, the laser beam should shine on a quadrant of the QPD. Using the two screws on the HPT1 cage mount, adjust the beam such that it shines on the center of the QPD. Congratulations, you have now aligned your optical trap! | ||
| + | |||
| + | ==Matlab OTKB GUI== | ||
| + | The manual that describes how to operate the Matlab OTKB GUI as well as describes the hardware setup and shows a picture of the GUI running is linked below. | ||
| + | |||
| + | [[Image:Matlab_OTKB_GUI_Manual.pdf]] | ||
==Comparison of OTKB and 20.309 optical trap operating characteristics== | ==Comparison of OTKB and 20.309 optical trap operating characteristics== | ||
| Line 100: | Line 83: | ||
|- | |- | ||
!X position | !X position | ||
| − | |[[Image:X- | + | |[[Image:X-axis_sensitivity_OTKB.gif|300px|X-axis Position Calibration]] |
| − | |[[Image:X- | + | |[[Image:X-axis_sensitivity.gif|300px|X-axis Position Calibration]] |
|- | |- | ||
!Y position | !Y position | ||
| − | | | + | |[[Image:Y-axis_sensitivity_OTKB.gif|300px|Y-axis Position Calibration]] |
| − | |[[Image:Y- | + | |[[Image:Y-axis_sensitivity.gif|300px|Y-axis Position Calibration]] |
|} | |} | ||
====Methodology==== | ====Methodology==== | ||
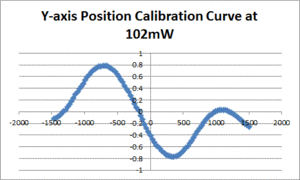
| − | [[Image: | + | [[Image:102mW_Position_Curve.gif|300px|thumb|Figure 1: A sample position calibration curve from the 20.309 trap]] |
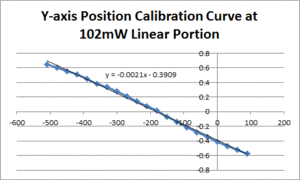
| − | [[Image: | + | [[Image:102mW_Position_Line.gif|300px|thumb|Figure 2: The linear portion of the curve from figure 1 fit to a line]] |
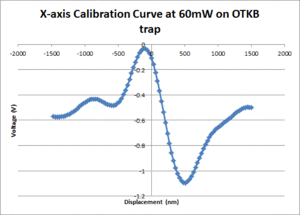
| + | [[Image:Calibration_curve_OTKB.gif|300px|thumb|Figure 3: A position calibration curve at 60mW on the OTKB trap]] | ||
#Prepare a sample cell per section 3.5 of the [http://scripts.mit.edu/~20.309/wiki/images/2/27/Optical_Trapping_Lab_Manual.pdf Optical Trapping Lab Manual]and load it with stuck and suspended silica microspheres as per section 4.1.1. | #Prepare a sample cell per section 3.5 of the [http://scripts.mit.edu/~20.309/wiki/images/2/27/Optical_Trapping_Lab_Manual.pdf Optical Trapping Lab Manual]and load it with stuck and suspended silica microspheres as per section 4.1.1. | ||
#Trap a suspended microsphere and adjust the focus to place it near the middle vertically of the sample cell. (Crash the trapped bead into the cover-slip and then raise it about half of the height of the cell, which is about 100 microns.) | #Trap a suspended microsphere and adjust the focus to place it near the middle vertically of the sample cell. (Crash the trapped bead into the cover-slip and then raise it about half of the height of the cell, which is about 100 microns.) | ||
| Line 134: | Line 118: | ||
#Estimated sensitivity is affected by focus. | #Estimated sensitivity is affected by focus. | ||
| + | ===Force Calibration=== | ||
====Trap stiffness by equipartition method==== | ====Trap stiffness by equipartition method==== | ||
=====Background===== | =====Background===== | ||
| Line 158: | Line 143: | ||
!20.309 | !20.309 | ||
|- | |- | ||
| − | !X | + | !X position |
| + | |TBD | ||
| + | |[[Image:X-axis_equipartition.gif|300px|X-axis Equipartition]] | ||
| + | |- | ||
| + | !Y position | ||
|TBD | |TBD | ||
| − | |[[Image: | + | |[[Image:Y-axis_Equipartition.gif|300px|Y-axis Equipartition]] |
|} | |} | ||
| Line 210: | Line 199: | ||
=====Results===== | =====Results===== | ||
| + | These data show that the OTKB trap is much stiffer than the 20.309 trap by a factor of 3 at low power levels (i.e. 25mW) and up to a factor of 4 at higher power levels (i.e. 125mW). Part of this large disparity in trap stiffness can be attributed to the difference in wavelength between the lasers used in the OTKB and 20.309 traps. The former uses a laser with a wavelength of 975nm whereas the latter uses a laser with a wavelength of 862nm. The 975nm laser is absorbed much more by water than the 862nm laser (by almost 1 order of magnitude). Thus, the trap created by the 975nm laser would be stiffer than that created by the 862nm laser. | ||
{| border="1" textalign="center" | {| border="1" textalign="center" | ||
| Line 218: | Line 208: | ||
|- | |- | ||
!X-axis Trap Stiffness | !X-axis Trap Stiffness | ||
| − | |[[Image: | + | |[[Image:OTKB_X-axis_Rolloff.gif|300px|X-axis PSD-rolloff Stiffness Calibration]] |
|[[Image:X-axis_PSD_rolloff.gif|300px|X-axis PSD-rolloff Stiffness Calibration]] | |[[Image:X-axis_PSD_rolloff.gif|300px|X-axis PSD-rolloff Stiffness Calibration]] | ||
|- | |- | ||
!Y-axis Trap Stiffness | !Y-axis Trap Stiffness | ||
| − | | | + | |[[Image:OTKB_Y-axis_Rolloff.gif|300px|X-axis PSD-rolloff Stiffness Calibration]] |
|TBD | |TBD | ||
|} | |} | ||
| Line 228: | Line 218: | ||
===DNA Tether Assay=== | ===DNA Tether Assay=== | ||
====Introduction==== | ====Introduction==== | ||
| − | The purpose of performing this assay was to use a simple biological measurement to compare the data obtained from the OTKB trap to the 20.309 trap as well as several documented results. | + | The purpose of performing this assay was to use a simple biological measurement to compare the data obtained from the OTKB trap to the 20.309 trap as well as several documented results. We used both traps to create a DNA-tether stretching curve, and from those data, calculate the tether length. The tether length can be calculated from the distance between the minimum and maximum of the tether curve from the following equation: L<sub>t</sub> = |X<sub>min</sub> - X<sub>max</sub>|- 2d where L<sub>t</sub> is the tether length, X<sub>min</sub> and X<sub>max</sub> are the displacement values of the minimum and maximum respectively, and d is the bead diameter. Futher information regarding this assay can be found in <ref name="Block">Wang, et. al. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1184516/pdf/biophysj00036-0355.pdf Stretching DNA with Optical Tweezers]. Biophysical Journal Volume 72 March 1997 1335-1346</ref> and section V of <ref name="Appleyard">Appleyard, et. al. Optical trapping for undergraduates[http://web.mit.edu/~langlab/Publications/Appleyard-etal(2007).pdf Optical trapping for undergraduates]. Am. J. Phys., Vol. 75, No. 1, January 2007</ref>. |
====Materials==== | ====Materials==== | ||
| Line 239: | Line 229: | ||
#Microscope Slide | #Microscope Slide | ||
#1<math>\mu</math>m diameter polyscience streptavin coated silica beads | #1<math>\mu</math>m diameter polyscience streptavin coated silica beads | ||
| − | #50<math>\mu</math>g/mL solution of anti-dig antibodies | + | #50<math>\mu</math>g/mL solution of anti-dig antibodies in PBS |
| − | #1ng/<math>\mu</math>L dilution of 3500 base pair DNA (for more information on production, read part B of | + | #1ng/<math>\mu</math>L dilution of 3500 base pair DNA in Casein(for more information on production, read section V part B of <ref name="Appleyard" />. |
| − | ====Procedure | + | ====Procedure==== |
| − | The procedure | + | The procedure was similar to references <ref name="Block" /> and <ref name="Appleyard" />. |
| − | #Create a solution of 1mg/mL casein using PBST as the solvent | + | #Create a solution of 1mg/mL casein using PBST as the solvent. |
#Using the syringe and filter, filter the casein solution into a new centrifuge tube. | #Using the syringe and filter, filter the casein solution into a new centrifuge tube. | ||
#Create a slide per the aforementioned method. Be sure to use an etched coverslip! | #Create a slide per the aforementioned method. Be sure to use an etched coverslip! | ||
| Line 261: | Line 251: | ||
#Flow 50<math>\mu</math>L of casein through the slide twice to rinse it out. | #Flow 50<math>\mu</math>L of casein through the slide twice to rinse it out. | ||
#Seal slide with vacuum grease or nail polish | #Seal slide with vacuum grease or nail polish | ||
| + | |||
| + | ====Data Collection==== | ||
| + | #Load the slide on the trap, and focus close to the coverslip. | ||
| + | #Slowly raise the focus until you see beads. | ||
| + | #Move in the x and y axes to find an isolated tether. You can recognize whether a bead is tethered by its movement: if it does not move in the z-axis (i.e. for a specific slide height, the bead does not move in and out of focus), it is a tether. There should be quite a few of them. | ||
| + | #Either run the Bead Alignment VI or center the bead in the trap by hand. You will know when the bead is centered by the voltage signal from the QPD or the phyical movement of the bead respectively to the two listed methods. For the first method, if the voltage signal from the axis you are not moving is constant, you are centered on that axis. For the second method, if it looks like you are pulling the tether at an angle, you need to adjust such that the bead is pulled in a straight line. | ||
| + | #Run the Position Calibration VI with a step size of 30 for 180 steps. Note, the tether might be pulled out of the trap, but this large displacement is necessary to collect the entire stretching curve. Save the data | ||
| + | #Repeat for multiple tethers. | ||
| + | |||
| + | ====Data Analysis==== | ||
| + | [[Image:DNA_tether_stretching_curve_20.309.gif|300px|thumb|Figure 4: A sample DNA-tether stretching curve]] | ||
| + | #Load the collected data into an analysis program such as excel or matlab | ||
| + | #Graph the voltage signal versus displacement, and pinpoint the maximum and minimum of the curve by looking at the graph and raw data. The graph should look similar to figure 4. | ||
| + | #Calculate the tether length | ||
| + | #Repeat for all measured tethers and compare to published results in the previously cited references <ref name="Block" /> and <ref name="Appleyard" />. | ||
| + | |||
| + | ====Results==== | ||
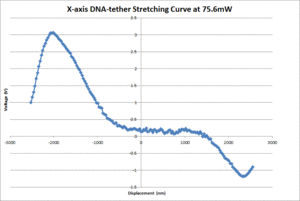
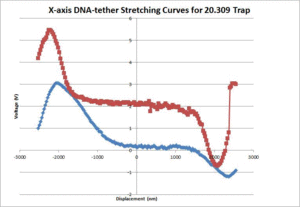
| + | [[Image:20.309_DNA-tether.gif|300px|thumb|Figure 5: X-axis 20.309 DNA-tether stretching curves]] | ||
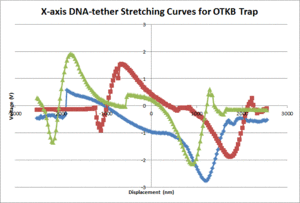
| + | [[Image:OTKB_DNA-tether.gif|300px|thumb|Figure 6: X-axis OTKB DNA-tether stretching curves]] | ||
| + | |||
| + | The average contour length of the DNA as measured by the 20.309 and OTKB traps were 1175nm and 335nm respectively. The 20.309 average countour length is nearly identical to the expected value of 1180nm stated in <ref name="Appleyard" /> whereas the OTKB value is off by about a factor of 3.5. We think this is mainly due to the centration method difference between the two traps. On the 20.309 trap, the joystick that controls the stage movement is much easier to use for centration than the fine adjustments on the thor stage or the APTUser software that controls the piezo cubes. We are in the middle of writing a VI for the OTKB trap that will center the tether in the trap and expect to see results similar to the 20.309 trap once that is completed and functional. The DNA-tether stretching curves that we have collected from both the 20.309 and the OTKB traps can be seen in figures 5 and 6 respectively. | ||
| + | |||
| + | ====Tips and techniques==== | ||
| + | *The casein solution should be made fresh daily. | ||
| + | *Casein is difficult to disolve. Spread it thinly on the inner wall of the centrifuge tube you use to make the solution. | ||
| + | |||
| + | ==References== | ||
| + | |||
| + | <references /> | ||
Latest revision as of 15:32, 5 August 2010
Contents
OTKB construction
Suggested modifications to OTKB layout
Note: numbers in parenthesis refer to the block diagram on page 4 of the Thorlabs "Optical Trap Application Setup" manual" or Thorlabs part numbers.
- The FiberPort Collimator (2) is connected to the Beam Steering Mirror (KCB1) with 4 cage rods (ER05), instead of a lens tube. (Use a 30mm optical cage plate gasket CPG3 to fill the small gap.)The FiberPort Collimator is no longer mounted on a cage plate mounting base (CPB1).
- The Beam Steering Mirror is connected to a 0.5" thick 30mm cage plate (CP02T) with 4 ER05 cage rods. Use a 0.3" SM1 lens tube (SM1L03) and an externally threaded tube coupler (SM1T2) to enclose the optical path.
- Use three 8" cage rods (ER8) rods to connect the CP02T to the 60mm cage cube (C6W) containing the Dichroic Mirror (4). Mount two 30mm cage plates (CP02) on the rods. The one closest to the KCB1 holds a 0.5' SM1 lens tube(SM1L05) that holds first achromatic doublet (AC254-060-B) and the second connected to a lens tube on either side and mounted on a base for stability. Plastic tubing encloses the optical path.
- A SM1L03 lens tube connected to a 1" SM1 lens tube (SM1L10) was screwed into the vertical segment's KCB1. The SM1L05, that holds the second achromatic doublet (part number AC-254-150-B), attached to the C6W, was connected to these combined lens tubes via a 1 inch piece of plastic tubing.
- The beam path in the vertical segment below the objective was enclosed by adding a SM1L05 lens tube connected to a SM1L03 lens tube threaded into it to the top of the KCB1. In addition, the cage translator (SM1Z) that originally held the objective was replaced by a CP02. This CP02 was mounted on the 4 3" cage rods (ER3) that were attached to the KCB1.
- The sample holder was changed from part AMA-SLH to a custom build slide holder attached to a mounting plate (RB13P1). We also reoriented the direction of the translating breadboard (TBB0606) so that it slides out parallel to the QPD segment.
- The height of the stage was lowered.
- Mount the(TBB0606) to the breadboard with four 2" long, 1.5" diameter mounting posts (P2). We eliminated the spacers (PS3) and bases (PB2) from each of the mounting legs.
Suggested modifications not yet implemented
- Change beam expander design from Keplerian to Galilean. Change lens 3A to p/n ACN254-040-B; change lens 3B to p/n AC254-075-B.
- Add a second cage cube for fluorescence imaging or a position detection laser.
- Use a separate tube lens, f=200mm, placed between the dichroic (4) and the CCD (5), instead of between the objective (6) and the dichroic (4).
- Implement a fine X/Y and height adjustment on the condenser. (The current implementation makes it difficult to center the laser in the condenser.)
- Implement a way to retract the condenser (Loading samples with the condenser in place is difficult.)
- Add a collimating/focusing lens to the LED illuminator. (This will reduce the image of the LED in the video capture and possibly facilitate easier condenser height adjustment.)
Alignment procedure
Note: throughout this procedure, the Relay Lens (3A) will be called the Beam Expander Lens and Relay Lens II (3B) will be called the Tube lens.
- Align and collimate the FiberPort as per section 4.1 of the ThorLabs Optical Trap Application Setup manual.
- Construct the beam expander segment and vertical segment before proceeding with the alignment.
- Mount the Beam Expander Lens (3A) and the Tube Lens (4A) in lens tubes, but set these aside instead of mounting the tubes in the cage plates. The position of the Beam Expander Lens will be adjusted by sliding the cage plate it is mounted to.
- Use 3 cage rods instead of 4 in the beam expander section so that lenses can be inserter and removed during the alignment procedure.
- Align the Beam Steering Mirror (4)
- Use two alignment jigs of your choosing (
Follow steps 9-12 in section 4.1 of the ThorLabs OTKB manual except don't attach the ring iris to the KCB1. Instead:
- Attach 3 ER8 cage rods to the KCB1.
- Slide 2 CP02s onto the cage rods, positioning one halfway to the end of the rods and the other at the end of the rods. Tighten the CP02s so they can't slide around.
- Attach an SM1D12D ring iris to each CP02 and open them all the way.
- Check your laser collimation and adjust the plunge screws accordingly to re-collimate your beam.
- Use the alignment techniques stated in the previous section to align the beam
- Remove the parts attached in steps 1-3.
Aligning the First Dichroic and Second KCB1
Complete up to step 10 of section 4.3 in the Thorlabs OTKB manual, skipping the alignment steps in section 4.2 (steps 21-22 and 30-31). It is useful to use 4 ER16 (2 ER8 rods connected together) rods instead of the 4 ER8 rods mentioned in step 11 of section 4.3. Then:
- Check the collimation of your beam by adjusting the CP02 holding the first achromatic doublet (f = 60mm). Secure this CP02 when you are confident in the laser collimation
- Close the iris from step 10 in section 4.3 of the OTKB manual. Adjust the rotate the dichroic and adjust its tilt screws so that the beam shoots through the center of the closed iris.
- Open that iris and close the iris mentioned in step 13 in section 4.3 of the OTKB manual. Using the 2 tilt screws on the KCB1, adjust the beam so that it shoots through the center of the closed iris.
- Repeat steps 2-3 until the beam goes through the center of both irises when they are closed.
- Remove the parts attached in steps 10-13 in section 4.3 of the OTKB manual
Collimating the Objective and Condenser
- Install the objective and the rest of the vertical segment as mentioned in section 4.3 of the OTKB manual accounting for the changes in the "Modifications" section.
- Mount the condenser as described in section 4.4 step 1-3 of the OTKB manual, disregarding the recommended height.
- Tighten the C1500 clamp holding the condenser such that it doesn't freely slide along the DP14 but it can still be moved very small amounts either by hand or by being tapped by a blunt instrument such as the handle of a screwdriver.
- Turn the laser on, load a sample slide (prepared as instructed in section 3.5 of the [1] using immersion oil, and adjust the condenser height by tapping on the C1500 clamp that holds it such that the beam is collimated. The condenser will be very close to the objective when properly collimated(on the order of a few mm).
- Tighten the C1500 clamp holding the condenser completely.
Aligning the Second Dichroic
- Complete section 4.4 of the OTKB manual (incorporating all necessary changes from the "Modifications" section)
- Attach the assembly mentioned in step 5 from the "Aligning the First Dichroic and Second KCB1" section to the face of the C6W to which the QPD will eventually be attached.
- Close the iris closest to the C6W and rotate the mount for the second dichroic such that the beam goes through the center of the closed iris.
- Open that iris and close the one furthest from the C6W. Adjust the tilt screws on the dichroic mount such that the beam goes through the center of the closed iris.
- Repeat steps 3-4 until the beam goes through the center of both irises when they are closed.
QPD
Assuming you have accurately aligned your trap, the laser beam should shine on a quadrant of the QPD. Using the two screws on the HPT1 cage mount, adjust the beam such that it shines on the center of the QPD. Congratulations, you have now aligned your optical trap!
Matlab OTKB GUI
The manual that describes how to operate the Matlab OTKB GUI as well as describes the hardware setup and shows a picture of the GUI running is linked below.
File:Matlab OTKB GUI Manual.pdf
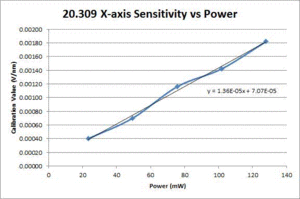
Comparison of OTKB and 20.309 optical trap operating characteristics
Position calibration
| ThorLabs OTKB | 20.309 | |
|---|---|---|
| X position | 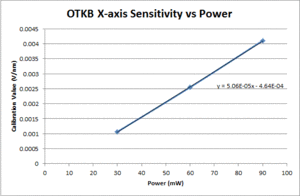
|
|
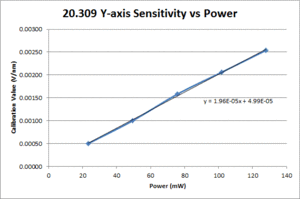
| Y position | 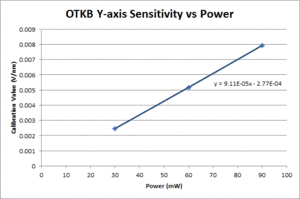
|
|
Methodology
- Prepare a sample cell per section 3.5 of the Optical Trapping Lab Manualand load it with stuck and suspended silica microspheres as per section 4.1.1.
- Trap a suspended microsphere and adjust the focus to place it near the middle vertically of the sample cell. (Crash the trapped bead into the cover-slip and then raise it about half of the height of the cell, which is about 100 microns.)
- Using QPD Alignment Tester, adjust the QPD position until the voltage output on both axes is near zero.
- Save an image of the trapped bead.
- Find a stuck bead.
- Adjust the focus so that the stuck bead looks similar to the saved image of the free trapped bead.
- Center the bead as per section 4.1.2. and run the Position Calibration VI. In the data file, the first row gives displacement from the starting position (in nm) and the second row gives the QPD signal for the axis being tested (in V).
- Fit a line to the approximately linear portion of the graph where the bead passes through the center of the trap. The slope of the line is the position sensitivity in V/nm.
- Repeat 5 times on 5 different stuck beads.
- Use the same procedure on the other axis
- Repeat the entire process at 5 power levels
- Compute the average sensitivity and standard deviation at each power level. Fit a line and plot to facilitate interpolation.
Materials
- 1μm silica microspheres, BangsLabs SS03N/4669
- 25x75mm glass slide
- 22x40mm #0 coverslip
- 1M NaCl
Comments
- Picomotor step size assumed to be 30nm. Using actual step size would increase accuracy of the position calibration.
- Estimated sensitivity is affected by focus.
Force Calibration
Trap stiffness by equipartition method
Background
Some background for this method can be found in section 4.2.1 of the Optical Trapping Lab Manual. Assuming these experiments are being run at room temperature, the equation from the lab manual simplifies to: $ \alpha = \frac{4.1124*10^-21}{\left \langle \Delta x^2 \right \rangle} $. Thus, in order to solve for the trap stiffness, the variance in position is the only thing that needs to be calculated. Since this variance is measured in volts, position calibrations will be required in order to convert the voltage signal into a position in meters (note: since the calibration values from the previous section were in V/nm, you will need to convert these into V/m in order to solve for $ \alpha $). The next section will explain the procedure used to perform this force calibration.
Procedure
- Prepare a sample cell per section 3.5 of the Optical Trapping Lab Manualand load it with about 10 μ; of 1:50,000 10% wt stock beads (1 μm silica, Bangs, SS03N/4669).
- Load the slide onto the microscope and trap a bead.
- Raise the bead to the middle of the channel and move it away from any obstructions.
- Use QPD Alignment Tester VI to center the voltage signal.
- Open the WriteXYTraceToFile VI. Enter the Sampling rate in Hz and the sampling time in seconds (64000 Hz and 3 sec were used respectively). Press start. While the VI is running, verify that the bead leaves the trap and nothing else becomes trapped. Save the data.
- Use the Matlab to calculate the variance of the position data. The result will be in m2.
- Use the sensitivity determined from position calibration to convert to meters2.
- Use the equation mentioned in the background section to calculate the trap stiffness.
- Repeat the calibration 10 times at each of 5 power levels (make sure to use different beads!) and average the stiffness results obtained per power level.
- Plot measured trap stiffness versus the power level and fit a line to facilitate interpolation.
Results
| ThorLabs OTKB | 20.309 | |
|---|---|---|
| X position | TBD | 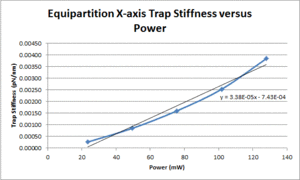
|
| Y position | TBD | 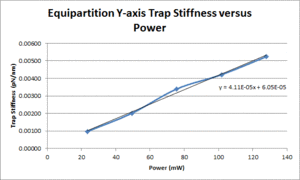
|
Trap stiffness by PSD rolloff method
Background
Some background for this method can be found in section 4.2.2 of the Optical Trapping Lab Manual. From the equations provided in the lab manual, we can solve for $ \alpha $ to be equal to: $ \alpha = 6\pi $2$ \eta $ d f0, which, plugging in for $ \eta $, $ \pi $, and d, we get that $ \alpha $ is directly proportional to f0, or the cutoff frequency, per the equation: $ \alpha $ = (5.27*10^-5)(f0), where $ \alpha $ is in pN/nm and f0 is in Hz. Below, the procedure for data collection and analysis as well as the matlab scripts used for the latter are written.
Procedure
- Load the collected waveform data from the equipartition stiffness calibrations for each trial into Matlab (i.e. data = load('filename');)
- Take a psd of the data, making use of the matlab command "pwelch" (i.e. datapsd = pwelch(data);)
- Create a frequency spectrum, f, that is based on the length of the psd by use of the matlab comand "linspace". If you sampled at 64kHz, you will want to type: f = linspace(1, 32000, 32769) where the inputs to the command "linspace" are the starting value, stopping value, and number of values respectively. Note: the sizes of f and the transpose of psd (i.e. psd') should be the same. If they aren't, you most likely entered the wrong values when creating f.
- Run the FitPSD(f, psd) Matlab function below to calculate the cut off frequency of the psd of the collected waveform data. Because of the 1/f noise that usually is present in the psd data, you may want to cut off the first 5-10 points when running this function (i.e. type FitPSD(f(10:end), psd(10:end)) to cut off the first 10 points). When you run the FitPSD function, it will return a 1x2 array. The number positioned at entry (1, 2) is the cutoff frequency. The function will also graph the raw PSD data as well as the nonlinear fit that it has attributed to the data. If the fit does not resemble the data well, cut off a few more of the beginning points. If it still doesn't fit the data, retake the data for that trial.
- Use the equation mentioned in the background section to calculate the trap stiffness, plugging in the cutoff frequency given by the matlab function mentioned in the previous step.
- Repeat for all collected data
- Plot measured trap stiffness versus the power level and fit a line to facilitate interpolation.
Analysis
Two matlab functions were used to analyze the waveform data collected. We made use of the matlab command "lsqcurvefit" to fit a nonlinear curve to our data based on the type of function which is documented in the lab manual referenced in the background portion of this section (for more information on how the matlab command "lsqcurvefit" works, check their documentation). The first matlab function is called "TransferFunc" and it takes two inputs: a 1x2 array called "params" and an array called "xdata". The two numbers in the array "params" are beta and the cutoff frequency (do not worry about the former). The second matlab function is called FitPSD and takes in two inputs: "f" and "psd", both of which you created in the previous section. The code for each of the functions is as follows (note, when saving each of these functions, make sure to save them as their function name as a .m file, so TransferFunc would be saved as TransferFunc.m. Otherwise, they will not work).:
TransferFunc.m:
function out = TransferFunc(params, xdata)
beta = params(1);
f0 = params(2);
out = params(1)./(params(2).^2 + xdata.^2);
FitPsd.m:
function out = FitPsd(f, psd)
out = lsqcurvefit(@TransferFunc, [1, 100], f, psd');
figure
loglog(f, psd)
hold on
loglog(f, TransferFunc(out, f), 'r');
Results
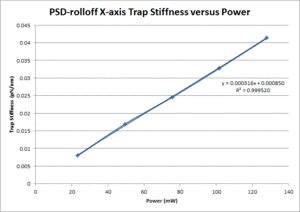
These data show that the OTKB trap is much stiffer than the 20.309 trap by a factor of 3 at low power levels (i.e. 25mW) and up to a factor of 4 at higher power levels (i.e. 125mW). Part of this large disparity in trap stiffness can be attributed to the difference in wavelength between the lasers used in the OTKB and 20.309 traps. The former uses a laser with a wavelength of 975nm whereas the latter uses a laser with a wavelength of 862nm. The 975nm laser is absorbed much more by water than the 862nm laser (by almost 1 order of magnitude). Thus, the trap created by the 975nm laser would be stiffer than that created by the 862nm laser.
| ThorLabs OTKB | 20.309 | |
|---|---|---|
| X-axis Trap Stiffness | 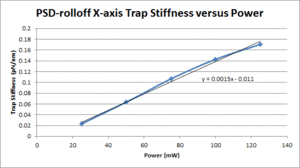
|
|
| Y-axis Trap Stiffness | 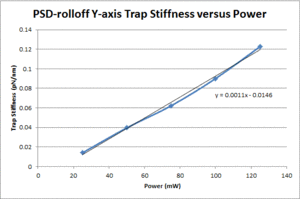
|
TBD |
DNA Tether Assay
Introduction
The purpose of performing this assay was to use a simple biological measurement to compare the data obtained from the OTKB trap to the 20.309 trap as well as several documented results. We used both traps to create a DNA-tether stretching curve, and from those data, calculate the tether length. The tether length can be calculated from the distance between the minimum and maximum of the tether curve from the following equation: Lt = |Xmin - Xmax|- 2d where Lt is the tether length, Xmin and Xmax are the displacement values of the minimum and maximum respectively, and d is the bead diameter. Futher information regarding this assay can be found in [1] and section V of [2].
Materials
- PBST
- PBS
- Casein
- Acrodisc Syringe Filter 0.2$ \mu $m HT Tuffryn Membrane
- BD 10ml Syringe
- Etched Coverslip
- Microscope Slide
- 1$ \mu $m diameter polyscience streptavin coated silica beads
- 50$ \mu $g/mL solution of anti-dig antibodies in PBS
- 1ng/$ \mu $L dilution of 3500 base pair DNA in Casein(for more information on production, read section V part B of [2].
Procedure
The procedure was similar to references [1] and [2].
- Create a solution of 1mg/mL casein using PBST as the solvent.
- Using the syringe and filter, filter the casein solution into a new centrifuge tube.
- Create a slide per the aforementioned method. Be sure to use an etched coverslip!
- Load 40$ \mu $L of the anti-dig solution onto the slide and let incubate coverslip down for 40 minutes.
- Pipet 5$ \mu $L of the streptavin coated silica beads into a microfuge tube.
- Add approximately 170$ \mu $L of PBST to the tube. Pipet up and down to suspend the beads
- Centrifuge the tube for 5 minutes at 10 rcf (10,000 rpm)
- Remove the tube from the centrifuge. You should notice a small pellet of beads at the bottom of the microfuge tube. Using a pipeter, remove the PBST from the microfuge tube being very careful as to not remove any beads.
- Repeat steps 6-8 two more times.
- Add 100$ \mu $L of the filtered casein solution to the microfuge tube holding the bead pellet, and pipet up and down to suspend the beads
- Vacuum 40$ \mu $L of the filtered casein solution into the slide channel and let sit for 30 minutes.
- Vacuum 40$ \mu $L of the DNA dilution into the slide channel and let sit for 30 minutes.
- Using a probe sonicator, sonicate the bead solution finished in step 10 for 2 minutes at a power level of 0.5W
- Vacuum 30$ \mu $L of the sonicated bead solution into the slide channel and let sit for 30 minutes
- Flow 50$ \mu $L of casein through the slide twice to rinse it out.
- Seal slide with vacuum grease or nail polish
Data Collection
- Load the slide on the trap, and focus close to the coverslip.
- Slowly raise the focus until you see beads.
- Move in the x and y axes to find an isolated tether. You can recognize whether a bead is tethered by its movement: if it does not move in the z-axis (i.e. for a specific slide height, the bead does not move in and out of focus), it is a tether. There should be quite a few of them.
- Either run the Bead Alignment VI or center the bead in the trap by hand. You will know when the bead is centered by the voltage signal from the QPD or the phyical movement of the bead respectively to the two listed methods. For the first method, if the voltage signal from the axis you are not moving is constant, you are centered on that axis. For the second method, if it looks like you are pulling the tether at an angle, you need to adjust such that the bead is pulled in a straight line.
- Run the Position Calibration VI with a step size of 30 for 180 steps. Note, the tether might be pulled out of the trap, but this large displacement is necessary to collect the entire stretching curve. Save the data
- Repeat for multiple tethers.
Data Analysis
- Load the collected data into an analysis program such as excel or matlab
- Graph the voltage signal versus displacement, and pinpoint the maximum and minimum of the curve by looking at the graph and raw data. The graph should look similar to figure 4.
- Calculate the tether length
- Repeat for all measured tethers and compare to published results in the previously cited references [1] and [2].
Results
The average contour length of the DNA as measured by the 20.309 and OTKB traps were 1175nm and 335nm respectively. The 20.309 average countour length is nearly identical to the expected value of 1180nm stated in [2] whereas the OTKB value is off by about a factor of 3.5. We think this is mainly due to the centration method difference between the two traps. On the 20.309 trap, the joystick that controls the stage movement is much easier to use for centration than the fine adjustments on the thor stage or the APTUser software that controls the piezo cubes. We are in the middle of writing a VI for the OTKB trap that will center the tether in the trap and expect to see results similar to the 20.309 trap once that is completed and functional. The DNA-tether stretching curves that we have collected from both the 20.309 and the OTKB traps can be seen in figures 5 and 6 respectively.
Tips and techniques
- The casein solution should be made fresh daily.
- Casein is difficult to disolve. Spread it thinly on the inner wall of the centrifuge tube you use to make the solution.
References
- ↑ 1.0 1.1 1.2 Wang, et. al. Stretching DNA with Optical Tweezers. Biophysical Journal Volume 72 March 1997 1335-1346
- ↑ 2.0 2.1 2.2 2.3 2.4 Appleyard, et. al. Optical trapping for undergraduatesOptical trapping for undergraduates. Am. J. Phys., Vol. 75, No. 1, January 2007