Difference between revisions of "MATLAB: Estimating resolution from a PSF slide image"
(→Measuring resolution) |
|||
| (19 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Category:20.309]] | ||
| + | [[Category:Matlab]] | ||
{{Template:20.309}} | {{Template:20.309}} | ||
| − | [[Image: | + | ===Measuring resolution=== |
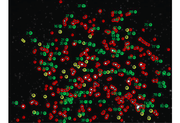
| + | [[Image:20.309_140218_PSFbeads.png|frameless|thumb|Example image processing on PSF beads to determine microscope resolution.]] | ||
| + | [[Image:20.309_140218_GaussianFit.png|frameless|thumb|Example Gaussian fit of a PSF bead fluorescence emission profile to estimate microscope resolution.]] | ||
| − | + | One of the most commonly used definitions of resolution is the distance between two point sources in the sample plane such that the ''peak'' of one point source’s ''blurred image'' falls on the ''first zero'' of the other source. The definition suggests a procedure for measuring resolution: make an image of a point source, measure the peak-to-trough distance in the image plane, and divide by the magnification. In this part of the lab, you will use a procedure inspired by this simple idea to estimate the resolution of your microscope. Instead of measuring the spot sizes with a ruler, you will use nonlinear regression to find best-fit parameters of digital images of point sources. | |
| − | + | One practical problem with this method is that true point sources are difficult to come by. If you are an astronomer using a telescope, stars are readily available, and they are very good approximations of point sources. Since there is no natural microscopic sample that is equivalent to the night sky, microscopists have to construct something suitable. One of the most common facsimiles is a microscope slide sprinkled with tiny, fluorescent beads that have diameters in the range of 100-190 nm. These beads are small enough to be considered point sources. Unfortunately, beads small enough for this purpose are not very bright. Imaging them can be challenging. Your microscope must be very well aligned to get good results. (The images you make for this part of the lab will probably remind you of the night sky. If they don't, have an instructor take a look at your setup.) | |
| − | + | In this part of the assignment, you will use image processing functions to locate the beads in an image and fit a Gaussian function to them. Gaussians are more amenable to nonlinear regression than Bessel functions, and they are a very good approximation. It is straightforward to convert the Gaussian parameters to Rayleigh resolution. See [[Converting Gaussian fit to Rayleigh resolution]] for a discussion of the conversion.[[Image:Synthetic PSF Image.png|thumb|right|Synthetic PSF image generated with MATLAB.]] | |
| − | The | + | ==Measuring resolution== |
| + | The sequence of steps for the resolution code is: | ||
| − | + | # find the connected regions | |
| − | function | + | # eliminate connected regions that are likely not image of a single microscphere |
| + | # use nonlinear regression to fit a Gaussian model function to the image | ||
| + | # compute summary statistics | ||
| + | # convert Gaussian parameter to Rayleigh resolution | ||
| − | + | ===Step 1: find the connected regions=== | |
| − | + | There are many methods for segmenting images. Fortunately, the fluorescent images of microspheres you are making in the lab usually have a high SNR. Because of this, a simple, global threshold works quite well. The bright pixels are the ones where the microspheres were. | |
| − | + | The function <tt> FindBrightObjectsInImage</tt> performs a global threshold and then computes various properties of the connected regions of pixels above the threshold. The MATLAB documentation includes [https://www.mathworks.com/help/images/ref/regionprops.html#inputarg_properties a complete list of the properties] that the built-in function <tt>regionprops</tt> can compute. | |
| − | + | <pre> | |
| + | function RegionProperties = FindBrightObjectsInImage( InputImage, GlobalThreshold, DilationRadius, PropertyList ) | ||
| − | + | mask = im2bw(InputImage, GlobalThreshold); | |
| + | mask = imclearborder(mask); % eliminates connected regions that touch the edge | ||
| + | mask = imdilate(mask, strel( 'disk', DilationRadius )); % open up the mask a little | ||
| − | + | RegionProperties = regionprops( mask, InputImage, PropertyList ); | |
| − | + | ||
end | end | ||
</pre> | </pre> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | The function below, <tt>EstimateResolutionFromPsfImage</tt>, takes an image of point sources (or approximate point sources) as an input and it returns an estimate of the resolution of an optical system that produced the image. It uses a global threshold to locate bright regions and the built-in MATLAB function <tt>[http://www.mathworks.com/help/images/ref/regionprops.html regionprops]</tt> to measure attributes of each connected region of bright pixels. You can add some code to the function that uses the properties of the regions to reject connected regions that are likely not images of single microspheres. The function uses the nonlinear regression function <tt>[http://www.mathworks.com/help/images/ref/nlinfit.html nlinfit]</tt> to estimate best fit Gaussian parameters for each bright spot. | |
| − | + | ||
| − | + | There are four subfunctions that should be included in the same m-file as EstimateResolutionFromPsfImage. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | |
| − | |||
| − | |||
<pre> | <pre> | ||
| − | function [ Resolution, StandardError, | + | function [ Resolution, StandardError, BestFitData ] = EstimateResolutionFromPsfImage( ImageData ) |
figure(1); | figure(1); | ||
| Line 65: | Line 56: | ||
% create bilevel image mapping bright regions | % create bilevel image mapping bright regions | ||
| − | thresholdLevel = 0.5 * ( max( ImageData(:) ) | + | thresholdLevel = 0.5 * ( max( ImageData(:) ) + median( ImageData(:) ) ); % this may not work in all cases |
binaryImage = im2bw( ImageData, thresholdLevel ); | binaryImage = im2bw( ImageData, thresholdLevel ); | ||
| − | dilatedImage = imdilate( binaryImage, strel( 'disk', 7, 0 ) ); | + | % increase the size of each connected region using dilation |
| − | dilatedImage = imclearborder( dilatedImage ); | + | dilatedImage = imdilate( binaryImage, strel( 'disk', 7, 0 ) ); |
| + | % remove objects touching the edges | ||
| + | dilatedImage = imclearborder( dilatedImage ); | ||
| − | + | % assign a unique object number to each connected region of pixels | |
| + | labeledImage = bwlabel( dilatedImage ); | ||
| − | CircleObjectsInImage( labeledImage, [ 1 0 0 ] ); | + | % draw a red circle around labeled objects |
| + | CircleObjectsInImage( labeledImage, [ 1 0 0 ] ); | ||
% compute the parameters of each object identified in the image | % compute the parameters of each object identified in the image | ||
| − | objectProperties = regionprops( dilatedImage, ImageData, 'Area', 'Centroid | + | objectProperties = regionprops( dilatedImage, ImageData, ... |
| + | 'Area', 'Centroid', 'PixelList', 'PixelValues', 'MaxIntensity' ); | ||
| − | % eliminate outliers -- touching, aggregates, etc... | + | % eliminate outliers -- PSF beads that are touching, aggregates, etc... |
| − | + | % begin by eliminating objects whose areas are outliers, as occurs when | |
| − | % | + | % beads are too close |
| − | % | + | medianArea = median( [ objectProperties.Area ] ); |
| − | + | outliers = [ objectProperties.Area ] > OutlierTolerancePercentage(2) * medianArea | [ objectProperties.Area ] < OutlierTolerancePercentage(1) * medianArea; | |
| − | outliers = [ objectProperties.Area ] > | + | |
| − | + | ||
| − | + | ||
if( sum( outliers ) > 0 ) | if( sum( outliers ) > 0 ) | ||
| + | dilatedImage = RemoveObjectsFromImage( dilatedImage, objectProperties( outliers ) ); | ||
objectProperties( outliers ) = []; % remove outliers | objectProperties( outliers ) = []; % remove outliers | ||
end | end | ||
| − | + | ||
| − | % eliminate intensity outliers | + | CircleObjectsInImage( dilatedImage, [ 1 1 0 ] ); |
| + | |||
| + | % next eliminate intensity outliers | ||
medianMaxIntensity = median( [ objectProperties.MaxIntensity ] ); | medianMaxIntensity = median( [ objectProperties.MaxIntensity ] ); | ||
| − | outliers = [ objectProperties.MaxIntensity ] > | + | outliers = [ objectProperties.MaxIntensity ] > OutlierTolerancePercentage(2) * medianMaxIntensity | [ objectProperties.MaxIntensity ] < OutlierTolerancePercentage(1) * medianMaxIntensity; |
| + | outliers = outliers | [ objectProperties.MaxIntensity ] > 0.995; | ||
| − | |||
| − | |||
if( sum( outliers ) > 0 ) | if( sum( outliers ) > 0 ) | ||
| + | dilatedImage = RemoveObjectsFromImage( dilatedImage, objectProperties( outliers ) ); | ||
objectProperties( outliers ) = []; | objectProperties( outliers ) = []; | ||
end | end | ||
| − | + | ||
| − | + | % circle all the remaining objects in green | |
| − | + | CircleObjectsInImage( dilatedImage, [ 0 1 0 ] ); | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | % | + | |
| − | CircleObjectsInImage( | + | |
LabelObjectsInImage( objectProperties ); | LabelObjectsInImage( objectProperties ); | ||
hold off; | hold off; | ||
| − | + | BestFitData = cell(1, length(objectProperties)); | |
% use nlinfit to fit a Gaussian to each object | % use nlinfit to fit a Gaussian to each object | ||
| Line 128: | Line 118: | ||
min(objectProperties(ii).PixelValues) ]; | min(objectProperties(ii).PixelValues) ]; | ||
| − | + | BestFitData{ii} = nlinfit( objectProperties(ii).PixelList, objectProperties(ii).PixelValues, @Gaussian2DFitFunction, initialGuesses ); | |
% plot data, initial guess, and fit for each peak | % plot data, initial guess, and fit for each peak | ||
figure(2) | figure(2) | ||
clf | clf | ||
| − | gd = delaunay( objectProperties(ii).PixelList(:,1), objectProperties(ii).PixelList(:,2) ); | + | |
| − | trimesh( gd, objectProperties(ii).PixelList(:,1), objectProperties(ii).PixelList(:,2), Gaussian2DFitFunction( | + | % generate a triangle mesh from the best fit solution found by |
| + | % nlinfit and plot it | ||
| + | gd = delaunay( objectProperties(ii).PixelList(:,1), ... | ||
| + | objectProperties(ii).PixelList(:,2) ); | ||
| + | trimesh( gd, objectProperties(ii).PixelList(:,1), ... | ||
| + | objectProperties(ii).PixelList(:,2), ... | ||
| + | Gaussian2DFitFunction(BestFitData{ii}, ... | ||
| + | objectProperties(ii).PixelList ) ) | ||
hold on | hold on | ||
| − | plot3( objectProperties(ii).PixelList(:,1), objectProperties(ii).PixelList(:,2),Gaussian2DFitFunction(initialGuesses, objectProperties(ii).PixelList ), 'rx' ) | + | |
| − | plot3( objectProperties(ii).PixelList(:,1), objectProperties(ii).PixelList(:,2), objectProperties(ii).PixelValues, 'gx', 'LineWidth', 3) | + | % plot initial guesses -- commented out to make plots less |
| + | % cluttered. put this back in to debug initial guesses | ||
| + | % plot3( objectProperties(ii).PixelList(:,1), ... | ||
| + | % objectProperties(ii).PixelList(:,2), ... | ||
| + | % Gaussian2DFitFunction(initialGuesses, ... | ||
| + | % objectProperties(ii).PixelList ), 'rx' ) | ||
| + | |||
| + | % plot image data | ||
| + | plot3( objectProperties(ii).PixelList(:,1), ... | ||
| + | objectProperties(ii).PixelList(:,2), ... | ||
| + | objectProperties(ii).PixelValues, 'gx', 'LineWidth', 3) | ||
| + | title(['Image data vs. Best Fit for Object Number ' num2str(ii)]); | ||
end | end | ||
| − | allPeakData = | + | allPeakData = vertcat( BestFitData{:} ); |
| − | Resolution = mean( allPeakData(:,4) ); | + | Resolution = mean( allPeakData(:,4) ) ./ 0.336; |
| − | StandardError = std( allPeakData(:,4) ) ./ sqrt( length( | + | StandardError = std( allPeakData(:,4) ./ 0.336 ) ./ sqrt( length( BestFitData ) ); |
end | end | ||
| Line 152: | Line 160: | ||
offset = Parameters(5); | offset = Parameters(5); | ||
| − | out = amplitude * exp( -(( Coordinates(:, 1) - yCenter ).^2 + ( Coordinates(:, 2) - xCenter ).^2 ) ./ (2 * sigma .^ 2 )) + offset; | + | out = amplitude * ... |
| + | exp( -(( Coordinates(:, 1) - yCenter ).^2 + ( Coordinates(:, 2) - xCenter ).^2 ) ... | ||
| + | ./ (2 * sigma .^ 2 )) + offset; | ||
end | end | ||
| Line 171: | Line 181: | ||
for ii = 1:length(ObjectProperties) | for ii = 1:length(ObjectProperties) | ||
unweightedCentroid = ObjectProperties(ii).Centroid; % Get centroid. | unweightedCentroid = ObjectProperties(ii).Centroid; % Get centroid. | ||
| − | text(unweightedCentroid(1) + labelShift, unweightedCentroid(2), num2str(ii), 'FontSize', fontSize, 'HorizontalAlignment', 'Right', 'Color', [0 1 0]); | + | text(unweightedCentroid(1) + labelShift, unweightedCentroid(2), ... |
| + | num2str(ii), 'FontSize', fontSize, 'HorizontalAlignment', ... | ||
| + | 'Right', 'Color', [0 1 0]); | ||
end | end | ||
end | end | ||
| + | |||
| + | function OutputBinaryImage = RemoveObjectsFromImage( InputBinaryImage, ObjectProperties ) | ||
| + | OutputBinaryImage = InputBinaryImage; | ||
| + | eliminatedPixels = vertcat( ObjectProperties.PixelList ); | ||
| + | allObjectIndexes = sub2ind( size( InputBinaryImage ), ... | ||
| + | eliminatedPixels(:, 2), eliminatedPixels(:,1) ); | ||
| + | OutputBinaryImage( allObjectIndexes ) = 0; | ||
| + | end | ||
| + | |||
| + | % initial version 9/23/2013 by SCW | ||
</pre> | </pre> | ||
| − | + | ==Testing the code== | |
| + | It is an excellent idea to test functions before using them. One way to test <tt>EstimateResolutionFromPsfImage</tt> is to generate a synthetic point-source image of known resolution. | ||
| − | + | ===Generating a synthetic PSF image=== | |
| − | + | <tt>SimulatePsfSlide</tt> repeatedly calls subfunction <tt>SimulateAiryDiskImage</tt> to generate a synthetic PSF slide image. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | <pre> | |
| + | function SimulatedImage = SimulatePsfSlide( ... | ||
NumericalAperture, Magnification, Wavelength, ... | NumericalAperture, Magnification, Wavelength, ... | ||
Intensity, PixelSize, ImageSize, NumberOfBeads, ... | Intensity, PixelSize, ImageSize, NumberOfBeads, ... | ||
| − | ProbabilityOfAggregation, TechnicalNoise, ElectronsPerCount ); | + | ProbabilityOfAggregation, TechnicalNoise, ElectronsPerCount ) |
| − | + | ||
| + | resolution = 0.61 * Wavelength / NumericalAperture; | ||
| + | airyDiskRadiusPixels = resolution * Magnification / PixelSize; | ||
| + | |||
| + | SimulatedImage = zeros( ImageSize ); | ||
| + | |||
| + | for ii = 1:NumberOfBeads | ||
| + | beadCenter = rand( 1, 2 ) .* ImageSize; | ||
| + | intensity = ( 1 + 0.1 * randn() ) * Intensity * ( 1 + poissrnd(ProbabilityOfAggregation) ); | ||
| + | SimulatedImage = SimulatedImage + ... | ||
| + | SimulateAiryDiskImage( beadCenter, airyDiskRadiusPixels, ImageSize, intensity ); | ||
| + | end | ||
| + | |||
| + | SimulatedImage = SimulatedImage + TechnicalNoise .* randn( ImageSize ); | ||
| + | SimulatedImage = uint16( round( SimulatedImage ./ ElectronsPerCount ) ); | ||
| + | end | ||
| + | |||
| + | function out = SimulateAiryDiskImage( AiryDiskCenter, AiryDiskRadius, ImageSize, Intensity ) | ||
| + | |||
| + | yAxis = (1:ImageSize(1)) - AiryDiskCenter(1); | ||
| + | xAxis = (1:ImageSize(2)) - AiryDiskCenter(2); | ||
| + | |||
| + | [ xCoordinate, yCoordinate ] = meshgrid( xAxis, yAxis ); | ||
| + | |||
| + | firstBesselZero = 3.8317; | ||
| + | normalizedRadius = firstBesselZero .* sqrt( xCoordinate.^2 + yCoordinate.^2 ) ./ AiryDiskRadius; | ||
| + | |||
| + | out = Intensity .* ( 2 * besselj(1, normalizedRadius ) ./ ( normalizedRadius ) ) .^ 2; | ||
| + | |||
| + | % remove NaN from result if center is an integer | ||
| + | if( all(AiryDiskCenter == fix( AiryDiskCenter )) ) | ||
| + | out( AiryDiskCenter(1), AiryDiskCenter(2) ) = Intensity; | ||
| + | end | ||
| + | end | ||
| − | |||
</pre> | </pre> | ||
| + | |||
| + | ===Testing the code=== | ||
| + | The script below generates synthetic images over a range of resolutions and compares them to the expected results. | ||
| + | [[Image:Code Test Results.png|thumb|right|Code testing results.]] | ||
| + | |||
| + | <pre> | ||
| + | |||
| + | naValues = 0.65:.025:1.25; | ||
| + | figureHandle = figure(); | ||
| + | |||
| + | measuredResolution = zeros( 1, length(naValues) ); | ||
| + | theoreticalResolution = zeros( 1, length(naValues) ); | ||
| + | standardError = zeros( 1, length(naValues) ); | ||
| + | |||
| + | for ii=1:length(naValues) | ||
| + | % generate synthetic PSF image | ||
| + | NumericalAperture = naValues(ii); %0.5; %65; | ||
| + | Magnification = 40; | ||
| + | Wavelength = 590e-9; % m | ||
| + | ProbabilityOfAggregation = 0; %0.2; | ||
| + | PixelSize = 7.4e-6; % m | ||
| + | ImageSize = [500 500]; % pixels x pixels | ||
| + | NumberOfBeads = 100; | ||
| + | TechnicalNoise = 0; % electrons per exposure | ||
| + | ElectronsPerCount = 2; | ||
| + | Intensity = 4095 * ElectronsPerCount * 0.8; % photons per exposure at peak | ||
| + | |||
| + | simulatedPsfImage = SimulatePsfSlide( ... | ||
| + | NumericalAperture, Magnification, Wavelength, ... | ||
| + | Intensity, PixelSize, ImageSize, NumberOfBeads, ... | ||
| + | ProbabilityOfAggregation, TechnicalNoise, ElectronsPerCount ); | ||
| + | |||
| + | % detect and simulate clipping due to overexposure | ||
| + | if( max( simulatedPsfImage(:) ) > 4095 ) | ||
| + | disp('*** Overexposed Image ***'); | ||
| + | simulatedPsfImage( simulatedPsfImage > 4095 ) = 4095; | ||
| + | end | ||
| + | |||
| + | simulatedPsfImage = im2double( simulatedPsfImage * 16 ); | ||
| + | |||
| + | theoreticalResolution(ii) = 0.61 * Wavelength / NumericalAperture; | ||
| + | |||
| + | [ measuredResolution(ii), standardError(ii), bestFitData ] = ... | ||
| + | EstimateResolutionFromPsfImage( simulatedPsfImage ); | ||
| + | |||
| + | measuredResolution(ii) = measuredResolution(ii) .* PixelSize / Magnification; | ||
| + | |||
| + | ==Effect of finite bead size == | ||
| + | Here is some code you can use to investigate the effect of finite bead size: | ||
| + | <pre> | ||
| + | close all | ||
| + | xAxis = -10:0.001:10; | ||
| + | |||
| + | % generate an Airy disk profile with a normalized resolution R=1 | ||
| + | firstBesselZero = 3.8317; | ||
| + | airyFunction = (2 * besselj(1, firstBesselZero * xAxis)./ (firstBesselZero * xAxis)).^2; | ||
| + | airyFunction(xAxis == 0) = 1; | ||
| + | |||
| + | % use nonlinear regression to fit a Gaussian function to the Airy disk | ||
| + | gaussianFunction = @(parameters, xdata) parameters(2) * exp( -xdata .^ 2 / (2 * parameters(1) .^ 2) ); | ||
| + | |||
| + | bestFitGaussianParameters = nlinfit(xAxis, airyFunction, gaussianFunction, [1 1]); | ||
| + | lsqcurvefit(gaussianFunction, [1 1], xAxis, airyFunction); | ||
| + | bestFitGaussian = gaussianFunction(bestFitGaussianParameters, xAxis); | ||
| + | sigma = bestFitGaussianParameters(1); | ||
| + | sigmaToFwhmConversionFactor = 2 * sqrt(2*log(2)); | ||
| + | fwhm = sigmaToFwhmConversionFactor * sigma; | ||
| + | |||
| + | % plot the results | ||
| + | plot(xAxis, airyFunction); | ||
| + | hold on; | ||
| + | plot(xAxis, bestFitGaussian, 'r'); | ||
| + | title(texlabel(['Airy Function (R=1) versus Gaussian (sigma = ' num2str(sigma) ' FWHM = ' num2str(fwhm) ')'])); | ||
| + | |||
| + | % generate a simulated bead with diameter = 0.33 * R | ||
| + | beadRadius = 0.33 / 2; | ||
| + | bead = ( abs( xAxis ) < beadRadius ) .* cos( pi / beadRadius / 2 * xAxis ); | ||
| + | bead = bead / sum( bead ); | ||
| + | beadConvolvedWithAiry = conv( airyFunction, bead, 'same' ); | ||
| + | |||
| + | figure | ||
| + | |||
| + | plot( xAxis, bead / max( bead ) ); | ||
| + | hold on | ||
| + | plot(xAxis, airyFunction, 'r'); | ||
| + | plot( xAxis, beadConvolvedWithAiry, 'g' ); | ||
| + | |||
| + | bestFitGaussianParameters = nlinfit(xAxis, beadConvolvedWithAiry, gaussianFunction, [1 1]); | ||
| + | bestFitGaussian = gaussianFunction(bestFitGaussianParameters, xAxis); | ||
| + | sigma = bestFitGaussianParameters(1); | ||
| + | sigmaToFwhmConversionFactor = 2 * sqrt(2*log(2)); | ||
| + | fwhm = sigmaToFwhmConversionFactor * sigma; | ||
| + | title(texlabel(['Airy Function Convolved with Bead Fit to Gaussian (sigma = ' num2str(sigma) ' FWHM = ' num2str(fwhm) ')'])); | ||
| + | </pre> | ||
| + | |||
| + | ===Making the simulation more realistic — adding noise.=== | ||
| + | The simulation is unrealistic in several ways. Add models for noise, optical aberrations, nonuniform illumination, etc... to characterize the effect of nonidealities on measurement accuracy and uncertainty. | ||
| + | |||
{{Template:20.309 bottom}} | {{Template:20.309 bottom}} | ||
Latest revision as of 14:21, 30 September 2017
Measuring resolution
One of the most commonly used definitions of resolution is the distance between two point sources in the sample plane such that the peak of one point source’s blurred image falls on the first zero of the other source. The definition suggests a procedure for measuring resolution: make an image of a point source, measure the peak-to-trough distance in the image plane, and divide by the magnification. In this part of the lab, you will use a procedure inspired by this simple idea to estimate the resolution of your microscope. Instead of measuring the spot sizes with a ruler, you will use nonlinear regression to find best-fit parameters of digital images of point sources.
One practical problem with this method is that true point sources are difficult to come by. If you are an astronomer using a telescope, stars are readily available, and they are very good approximations of point sources. Since there is no natural microscopic sample that is equivalent to the night sky, microscopists have to construct something suitable. One of the most common facsimiles is a microscope slide sprinkled with tiny, fluorescent beads that have diameters in the range of 100-190 nm. These beads are small enough to be considered point sources. Unfortunately, beads small enough for this purpose are not very bright. Imaging them can be challenging. Your microscope must be very well aligned to get good results. (The images you make for this part of the lab will probably remind you of the night sky. If they don't, have an instructor take a look at your setup.)
In this part of the assignment, you will use image processing functions to locate the beads in an image and fit a Gaussian function to them. Gaussians are more amenable to nonlinear regression than Bessel functions, and they are a very good approximation. It is straightforward to convert the Gaussian parameters to Rayleigh resolution. See Converting Gaussian fit to Rayleigh resolution for a discussion of the conversion.Measuring resolution
The sequence of steps for the resolution code is:
- find the connected regions
- eliminate connected regions that are likely not image of a single microscphere
- use nonlinear regression to fit a Gaussian model function to the image
- compute summary statistics
- convert Gaussian parameter to Rayleigh resolution
Step 1: find the connected regions
There are many methods for segmenting images. Fortunately, the fluorescent images of microspheres you are making in the lab usually have a high SNR. Because of this, a simple, global threshold works quite well. The bright pixels are the ones where the microspheres were.
The function FindBrightObjectsInImage performs a global threshold and then computes various properties of the connected regions of pixels above the threshold. The MATLAB documentation includes a complete list of the properties that the built-in function regionprops can compute.
function RegionProperties = FindBrightObjectsInImage( InputImage, GlobalThreshold, DilationRadius, PropertyList )
mask = im2bw(InputImage, GlobalThreshold);
mask = imclearborder(mask); % eliminates connected regions that touch the edge
mask = imdilate(mask, strel( 'disk', DilationRadius )); % open up the mask a little
RegionProperties = regionprops( mask, InputImage, PropertyList );
end
The function below, EstimateResolutionFromPsfImage, takes an image of point sources (or approximate point sources) as an input and it returns an estimate of the resolution of an optical system that produced the image. It uses a global threshold to locate bright regions and the built-in MATLAB function regionprops to measure attributes of each connected region of bright pixels. You can add some code to the function that uses the properties of the regions to reject connected regions that are likely not images of single microspheres. The function uses the nonlinear regression function nlinfit to estimate best fit Gaussian parameters for each bright spot.
There are four subfunctions that should be included in the same m-file as EstimateResolutionFromPsfImage.
function [ Resolution, StandardError, BestFitData ] = EstimateResolutionFromPsfImage( ImageData )
figure(1);
imshow( ImageData );
hold on;
title( 'PSF Image' );
% create bilevel image mapping bright regions
thresholdLevel = 0.5 * ( max( ImageData(:) ) + median( ImageData(:) ) ); % this may not work in all cases
binaryImage = im2bw( ImageData, thresholdLevel );
% increase the size of each connected region using dilation
dilatedImage = imdilate( binaryImage, strel( 'disk', 7, 0 ) );
% remove objects touching the edges
dilatedImage = imclearborder( dilatedImage );
% assign a unique object number to each connected region of pixels
labeledImage = bwlabel( dilatedImage );
% draw a red circle around labeled objects
CircleObjectsInImage( labeledImage, [ 1 0 0 ] );
% compute the parameters of each object identified in the image
objectProperties = regionprops( dilatedImage, ImageData, ...
'Area', 'Centroid', 'PixelList', 'PixelValues', 'MaxIntensity' );
% eliminate outliers -- PSF beads that are touching, aggregates, etc...
% begin by eliminating objects whose areas are outliers, as occurs when
% beads are too close
medianArea = median( [ objectProperties.Area ] );
outliers = [ objectProperties.Area ] > OutlierTolerancePercentage(2) * medianArea | [ objectProperties.Area ] < OutlierTolerancePercentage(1) * medianArea;
if( sum( outliers ) > 0 )
dilatedImage = RemoveObjectsFromImage( dilatedImage, objectProperties( outliers ) );
objectProperties( outliers ) = []; % remove outliers
end
CircleObjectsInImage( dilatedImage, [ 1 1 0 ] );
% next eliminate intensity outliers
medianMaxIntensity = median( [ objectProperties.MaxIntensity ] );
outliers = [ objectProperties.MaxIntensity ] > OutlierTolerancePercentage(2) * medianMaxIntensity | [ objectProperties.MaxIntensity ] < OutlierTolerancePercentage(1) * medianMaxIntensity;
outliers = outliers | [ objectProperties.MaxIntensity ] > 0.995;
if( sum( outliers ) > 0 )
dilatedImage = RemoveObjectsFromImage( dilatedImage, objectProperties( outliers ) );
objectProperties( outliers ) = [];
end
% circle all the remaining objects in green
CircleObjectsInImage( dilatedImage, [ 0 1 0 ] );
LabelObjectsInImage( objectProperties );
hold off;
BestFitData = cell(1, length(objectProperties));
% use nlinfit to fit a Gaussian to each object
for ii = 1:length(objectProperties)
% initial guess for sigma based on area of bright spot
maximumPixelValue = max( objectProperties(ii).PixelValues );
darkPixelValue = median( objectProperties(ii).PixelValues );
pixelCountAboveHalf = sum( objectProperties(ii).PixelValues > .5 * ( maximumPixelValue + darkPixelValue ) );
sigmaInitialGuess = 0.8 * sqrt( pixelCountAboveHalf / 2 / pi / log(2) );
initialGuesses = [ ...
objectProperties(ii).Centroid(1), ... % yCenter
objectProperties(ii).Centroid(2), ... % xCenter
max(objectProperties(ii).PixelValues) - min(objectProperties(ii).PixelValues), ... % amplitude
sigmaInitialGuess, ... % (objectProperties(ii).BoundingBox(3) - 6) / 4, ... % sigma
min(objectProperties(ii).PixelValues) ];
BestFitData{ii} = nlinfit( objectProperties(ii).PixelList, objectProperties(ii).PixelValues, @Gaussian2DFitFunction, initialGuesses );
% plot data, initial guess, and fit for each peak
figure(2)
clf
% generate a triangle mesh from the best fit solution found by
% nlinfit and plot it
gd = delaunay( objectProperties(ii).PixelList(:,1), ...
objectProperties(ii).PixelList(:,2) );
trimesh( gd, objectProperties(ii).PixelList(:,1), ...
objectProperties(ii).PixelList(:,2), ...
Gaussian2DFitFunction(BestFitData{ii}, ...
objectProperties(ii).PixelList ) )
hold on
% plot initial guesses -- commented out to make plots less
% cluttered. put this back in to debug initial guesses
% plot3( objectProperties(ii).PixelList(:,1), ...
% objectProperties(ii).PixelList(:,2), ...
% Gaussian2DFitFunction(initialGuesses, ...
% objectProperties(ii).PixelList ), 'rx' )
% plot image data
plot3( objectProperties(ii).PixelList(:,1), ...
objectProperties(ii).PixelList(:,2), ...
objectProperties(ii).PixelValues, 'gx', 'LineWidth', 3)
title(['Image data vs. Best Fit for Object Number ' num2str(ii)]);
end
allPeakData = vertcat( BestFitData{:} );
Resolution = mean( allPeakData(:,4) ) ./ 0.336;
StandardError = std( allPeakData(:,4) ./ 0.336 ) ./ sqrt( length( BestFitData ) );
end
function out = Gaussian2DFitFunction( Parameters, Coordinates )
yCenter = Parameters(1);
xCenter = Parameters(2);
amplitude = Parameters(3);
sigma = Parameters(4);
offset = Parameters(5);
out = amplitude * ...
exp( -(( Coordinates(:, 1) - yCenter ).^2 + ( Coordinates(:, 2) - xCenter ).^2 ) ...
./ (2 * sigma .^ 2 )) + offset;
end
function CircleObjectsInImage( LabelImage, BorderColor )
boundaries = bwboundaries( LabelImage );
numberOfBoundaries = size( boundaries );
for k = 1 : numberOfBoundaries
thisBoundary = boundaries{k};
plot(thisBoundary(:,2), thisBoundary(:,1), 'Color', BorderColor, 'LineWidth', 2);
end
end
function LabelObjectsInImage( ObjectProperties )
labelShift = -9;
fontSize = 10;
for ii = 1:length(ObjectProperties)
unweightedCentroid = ObjectProperties(ii).Centroid; % Get centroid.
text(unweightedCentroid(1) + labelShift, unweightedCentroid(2), ...
num2str(ii), 'FontSize', fontSize, 'HorizontalAlignment', ...
'Right', 'Color', [0 1 0]);
end
end
function OutputBinaryImage = RemoveObjectsFromImage( InputBinaryImage, ObjectProperties )
OutputBinaryImage = InputBinaryImage;
eliminatedPixels = vertcat( ObjectProperties.PixelList );
allObjectIndexes = sub2ind( size( InputBinaryImage ), ...
eliminatedPixels(:, 2), eliminatedPixels(:,1) );
OutputBinaryImage( allObjectIndexes ) = 0;
end
% initial version 9/23/2013 by SCW
Testing the code
It is an excellent idea to test functions before using them. One way to test EstimateResolutionFromPsfImage is to generate a synthetic point-source image of known resolution.
Generating a synthetic PSF image
SimulatePsfSlide repeatedly calls subfunction SimulateAiryDiskImage to generate a synthetic PSF slide image.
function SimulatedImage = SimulatePsfSlide( ...
NumericalAperture, Magnification, Wavelength, ...
Intensity, PixelSize, ImageSize, NumberOfBeads, ...
ProbabilityOfAggregation, TechnicalNoise, ElectronsPerCount )
resolution = 0.61 * Wavelength / NumericalAperture;
airyDiskRadiusPixels = resolution * Magnification / PixelSize;
SimulatedImage = zeros( ImageSize );
for ii = 1:NumberOfBeads
beadCenter = rand( 1, 2 ) .* ImageSize;
intensity = ( 1 + 0.1 * randn() ) * Intensity * ( 1 + poissrnd(ProbabilityOfAggregation) );
SimulatedImage = SimulatedImage + ...
SimulateAiryDiskImage( beadCenter, airyDiskRadiusPixels, ImageSize, intensity );
end
SimulatedImage = SimulatedImage + TechnicalNoise .* randn( ImageSize );
SimulatedImage = uint16( round( SimulatedImage ./ ElectronsPerCount ) );
end
function out = SimulateAiryDiskImage( AiryDiskCenter, AiryDiskRadius, ImageSize, Intensity )
yAxis = (1:ImageSize(1)) - AiryDiskCenter(1);
xAxis = (1:ImageSize(2)) - AiryDiskCenter(2);
[ xCoordinate, yCoordinate ] = meshgrid( xAxis, yAxis );
firstBesselZero = 3.8317;
normalizedRadius = firstBesselZero .* sqrt( xCoordinate.^2 + yCoordinate.^2 ) ./ AiryDiskRadius;
out = Intensity .* ( 2 * besselj(1, normalizedRadius ) ./ ( normalizedRadius ) ) .^ 2;
% remove NaN from result if center is an integer
if( all(AiryDiskCenter == fix( AiryDiskCenter )) )
out( AiryDiskCenter(1), AiryDiskCenter(2) ) = Intensity;
end
end
Testing the code
The script below generates synthetic images over a range of resolutions and compares them to the expected results.
naValues = 0.65:.025:1.25;
figureHandle = figure();
measuredResolution = zeros( 1, length(naValues) );
theoreticalResolution = zeros( 1, length(naValues) );
standardError = zeros( 1, length(naValues) );
for ii=1:length(naValues)
% generate synthetic PSF image
NumericalAperture = naValues(ii); %0.5; %65;
Magnification = 40;
Wavelength = 590e-9; % m
ProbabilityOfAggregation = 0; %0.2;
PixelSize = 7.4e-6; % m
ImageSize = [500 500]; % pixels x pixels
NumberOfBeads = 100;
TechnicalNoise = 0; % electrons per exposure
ElectronsPerCount = 2;
Intensity = 4095 * ElectronsPerCount * 0.8; % photons per exposure at peak
simulatedPsfImage = SimulatePsfSlide( ...
NumericalAperture, Magnification, Wavelength, ...
Intensity, PixelSize, ImageSize, NumberOfBeads, ...
ProbabilityOfAggregation, TechnicalNoise, ElectronsPerCount );
% detect and simulate clipping due to overexposure
if( max( simulatedPsfImage(:) ) > 4095 )
disp('*** Overexposed Image ***');
simulatedPsfImage( simulatedPsfImage > 4095 ) = 4095;
end
simulatedPsfImage = im2double( simulatedPsfImage * 16 );
theoreticalResolution(ii) = 0.61 * Wavelength / NumericalAperture;
[ measuredResolution(ii), standardError(ii), bestFitData ] = ...
EstimateResolutionFromPsfImage( simulatedPsfImage );
measuredResolution(ii) = measuredResolution(ii) .* PixelSize / Magnification;
==Effect of finite bead size ==
Here is some code you can use to investigate the effect of finite bead size:
<pre>
close all
xAxis = -10:0.001:10;
% generate an Airy disk profile with a normalized resolution R=1
firstBesselZero = 3.8317;
airyFunction = (2 * besselj(1, firstBesselZero * xAxis)./ (firstBesselZero * xAxis)).^2;
airyFunction(xAxis == 0) = 1;
% use nonlinear regression to fit a Gaussian function to the Airy disk
gaussianFunction = @(parameters, xdata) parameters(2) * exp( -xdata .^ 2 / (2 * parameters(1) .^ 2) );
bestFitGaussianParameters = nlinfit(xAxis, airyFunction, gaussianFunction, [1 1]);
lsqcurvefit(gaussianFunction, [1 1], xAxis, airyFunction);
bestFitGaussian = gaussianFunction(bestFitGaussianParameters, xAxis);
sigma = bestFitGaussianParameters(1);
sigmaToFwhmConversionFactor = 2 * sqrt(2*log(2));
fwhm = sigmaToFwhmConversionFactor * sigma;
% plot the results
plot(xAxis, airyFunction);
hold on;
plot(xAxis, bestFitGaussian, 'r');
title(texlabel(['Airy Function (R=1) versus Gaussian (sigma = ' num2str(sigma) ' FWHM = ' num2str(fwhm) ')']));
% generate a simulated bead with diameter = 0.33 * R
beadRadius = 0.33 / 2;
bead = ( abs( xAxis ) < beadRadius ) .* cos( pi / beadRadius / 2 * xAxis );
bead = bead / sum( bead );
beadConvolvedWithAiry = conv( airyFunction, bead, 'same' );
figure
plot( xAxis, bead / max( bead ) );
hold on
plot(xAxis, airyFunction, 'r');
plot( xAxis, beadConvolvedWithAiry, 'g' );
bestFitGaussianParameters = nlinfit(xAxis, beadConvolvedWithAiry, gaussianFunction, [1 1]);
bestFitGaussian = gaussianFunction(bestFitGaussianParameters, xAxis);
sigma = bestFitGaussianParameters(1);
sigmaToFwhmConversionFactor = 2 * sqrt(2*log(2));
fwhm = sigmaToFwhmConversionFactor * sigma;
title(texlabel(['Airy Function Convolved with Bead Fit to Gaussian (sigma = ' num2str(sigma) ' FWHM = ' num2str(fwhm) ')']));
Making the simulation more realistic — adding noise.
The simulation is unrealistic in several ways. Add models for noise, optical aberrations, nonuniform illumination, etc... to characterize the effect of nonidealities on measurement accuracy and uncertainty.