Difference between revisions of "Lab Manual: Measuring DNA Melting Curves"
Steven Nagle (Talk | contribs) (→Overview) |
Steven Nagle (Talk | contribs) (→Overview) |
||
| Line 32: | Line 32: | ||
A blue LED excites the sample. The LED produces a broad spectrum of light with a peak intensity around 475 nm. A band pass optical filter eliminates wavelengths below 450 and above 490 nm. | A blue LED excites the sample. The LED produces a broad spectrum of light with a peak intensity around 475 nm. A band pass optical filter eliminates wavelengths below 450 and above 490 nm. | ||
| − | A photodiode will be placed at 90° to the LED source to detect the green light emitted by SYBR Green I. | + | A photodiode will be placed at 90° to the LED source to detect the green light emitted by SYBR Green I. A long-pass emission filter will be placed between the sample and detector to pass only wavelengths longer than 515 nm. This filter, in combination with the LED band pass filter, eliminates any background signal due to the excitation light. |
A metal block will hold a sample vial in the excitation light path and transfer heat to the sample. The sample contains a combination of DNA, SYBR Green I and a salt. Recall that SYBR Green I fluoresces when it is bound to double stranded DNA (dsDNA) by a factor of up to 1000x compared to when it is bound to single stranded DNA (ssDNA). Detection of this fluorescence provides a relative measure of the fraction of dsDNA to total DNA, which can be compared to predictions from a thermodynamic model. | A metal block will hold a sample vial in the excitation light path and transfer heat to the sample. The sample contains a combination of DNA, SYBR Green I and a salt. Recall that SYBR Green I fluoresces when it is bound to double stranded DNA (dsDNA) by a factor of up to 1000x compared to when it is bound to single stranded DNA (ssDNA). Detection of this fluorescence provides a relative measure of the fraction of dsDNA to total DNA, which can be compared to predictions from a thermodynamic model. | ||
Revision as of 03:04, 24 April 2015
|
|
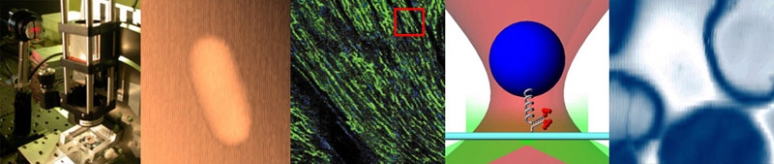
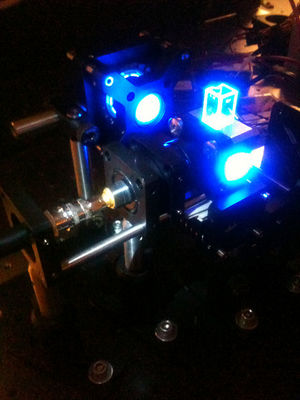
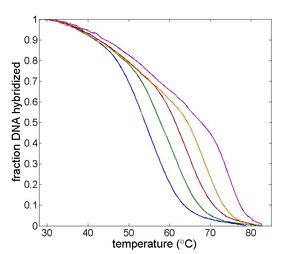
| Temperature cycling fluorometer apparatus. | Example DNA melting curves showing the effect of varying ionic strength. |
Overview
Complementary DNA oligos in solution exist in a temperature dependent equilibrium that consists predominantly of double stranded helicies and single stranded random coils. Double stranded DNA (dsDNA) predominates at low temperatures. As temperature increases, the equilibrium shifts toward single stranded DNA (ssDNA). The equilibrium may be characterized by a DNA melting curve, which is a plot of dsDNA concentration versus temperature. The shape of the melting curve depends on several characteristics of the oligos and the solution, including the length of the oligos; concentration of salt ions in solution; AT versus GC content; and the degree of complementarity of the oligos. Several web-based tools are available to predict melting melting curves, including DINAMelt Web Server and Oligocalc. A review of DNA melting thermodynamics is available on this wiki page.
In this lab, you will build an instrument called a temperature cycling fluorometer and use it to generate DNA melting curves for several oligos. The technique you will use exploits a fluorescent dye that glows brightly when bound to dsDNA, but hardly at all otherwise. Melting curves are recorded by measuring the intensity of green fluorescence coming from a sample excited by blue light as it is heated and cooled over a range of temperatures. Raw data from the instrument consists of two voltages: one proportional to fluorescence, and another that depends on temperature. You will digitize the voltages with a computer data acquisition system and process the raw signals to produce DNA melting curves. From the melting curves, you will estimate the thermodynamic parameters of the DNA annealing reaction: ΔH°, ΔS° and the melting temperature. You will use your instrument to examine the effects of oligo length, ion concentration, or degree of complementarity on the thermodynamic parameters and to identify an unknown sample.
The instrument has six major subsystems:
- sample
- optical excitation
- temperature control
- fluorescence detection
- temperature measurement
- data acquisition and control
The sample consists of 30 μM complementary DNA oligos in solution with NaCl and a fluorescent dye. Several dsDNA dyes are available with blue excitation and green emission , including [SYBR Green] and Resolight. SYBR Green has a peak sensitivity to blue light at 497 nm. The dye emits green light with an emission peak at 520 nm.
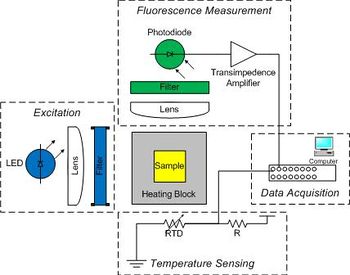
A blue LED excites the sample. The LED produces a broad spectrum of light with a peak intensity around 475 nm. A band pass optical filter eliminates wavelengths below 450 and above 490 nm.
A photodiode will be placed at 90° to the LED source to detect the green light emitted by SYBR Green I. A long-pass emission filter will be placed between the sample and detector to pass only wavelengths longer than 515 nm. This filter, in combination with the LED band pass filter, eliminates any background signal due to the excitation light.
A metal block will hold a sample vial in the excitation light path and transfer heat to the sample. The sample contains a combination of DNA, SYBR Green I and a salt. Recall that SYBR Green I fluoresces when it is bound to double stranded DNA (dsDNA) by a factor of up to 1000x compared to when it is bound to single stranded DNA (ssDNA). Detection of this fluorescence provides a relative measure of the fraction of dsDNA to total DNA, which can be compared to predictions from a thermodynamic model.
The sample block will be heated by a TEC (thermoelectric cooler) and its temperature measured by an embedded RTD (resistance temperature detector). Also, since photodiodes produce a very small amount of current, a high gain amplifier will convert the photodiode current into a measurable voltage. The DAQ (data acquisition card) and PC will digitize the amplified photodiode and RTD signals. A Matlab program is provided to record these fluorescence and temperature signals over time and save the data to a file. The file can then be loaded into Matlab for analysis, or if you prefer, Python or equivalent where the signals will be converted into dsDNA concentration and sample temperature. Finally, the dsDNA sample melting temperature will be estimated from a plot of df/dT vs T (f represents the fraction of dsDNA) and from a comparison to the thermodynamic model. The thermodynamic parameters of DNA melting depend on the sequence length, salt ion concentration, and degree of complementary between the two oligos. The measurement technique utilizes a fluorescent dye that binds preferentially to double stranded DNA (dsDNA). This characteristic of the dye allows the relative concentration of dsDNA to be determined by measuring the intensity of fluorescent light emitted by an excited sample.
You will measure samples of both known and unknown composition. The samples may vary in length, complementarity (complete match, single mismatch, or complete mismatch), or salt concentration. You will compare the data you gather to a theoretical model and you will attempt to identify unknown samples.
A common application of this technique exploits the length dependence of DNA melting temperatures to examine PCR products in order to determine whether a desired sequence was successfully amplified.
How to do this lab
- Refresh your understanding of DNA Melting Thermodynamics.
- Complete the Simulating DNA Melting homework using DNA Melting: Simulating DNA Melting - Basics and learn more about the signals you will observe in the lab and how to start to analyze them.
- Follow the guidelines in Part 1 of the lab to build a system for exciting, heating, measuring fluorescence, and measuring temperature of a DNA sample.
- Troubleshoot and optimize your instrument.
- Measure the signal to noise ratio.
- Generate melting curves for a known sample.
- Estimate the melting temperature of the known sample. Turn in Part 1 of your lab report.
- Improve your simulation of DNA Melting results by adding the modeled effects described in DNA Melting: Model function and parameter estimation by nonlinear regression.
- Improve your instrument by adding temperature control and lock-in signal processing, as outlined in Part 2 of the lab.
- Verify the performance of your instrument with known samples.
- Measure your unknown samples.
- Attend the tutorial on multi-parameter, nonlinear regression to further improve your model toward even better estimates of ΔH°, and ΔS°, and use to analyze your data from Part 2. Your goal is to identify the unknown sample.
- Turn in Part 2 of your lab report. In this final report submission include Part 2 only, but note any significant revisions to hardware, software or methods that you may have made since Part 1.
Objectives and learning goals
- Build an optical system for exciting the sample with blue light and gathering the fluorescence output on the photodiode.
- Measure light intensity with a photodiode.
- Build a heating system to reliably heat and cool your sample.
- Measure temperature with an RTD and an appropriate transfer function.
- Implement a high gain transimpedance amplifier.
- Use a lock-in amplifier to reduce noise.
- Record dsDNA concentration versus temperature curves for several samples.
- Analyze the data to find the dsDNA fraction as a function of temperature.
- Estimate Tm from your data.
- Compare the measured curves with theoretical models.
- Identify unknown DNA samples.
Lab manual sections
- Lab Manual:Measuring DNA Melting Curves
- DNA Melting: Simulating DNA Melting - Basics
- DNA Melting Part 1: Measuring Temperature and Fluorescence
- DNA Melting Part 2: Lock-in Amplifier and Temperature Control
Suggested readings and references
A more complete DNA melting and PCR resource list is available on this wiki. Please improve the page by adding relevant, high-quality sources.