Lab 1 Report -- Nathan S Lachenmyer
From Course Wiki
Revision as of 06:15, 12 March 2012 by Nathan S Lachenmyer (Talk | contribs)
Contents
Calibration
All calibrations and experiments done at 20 mW.
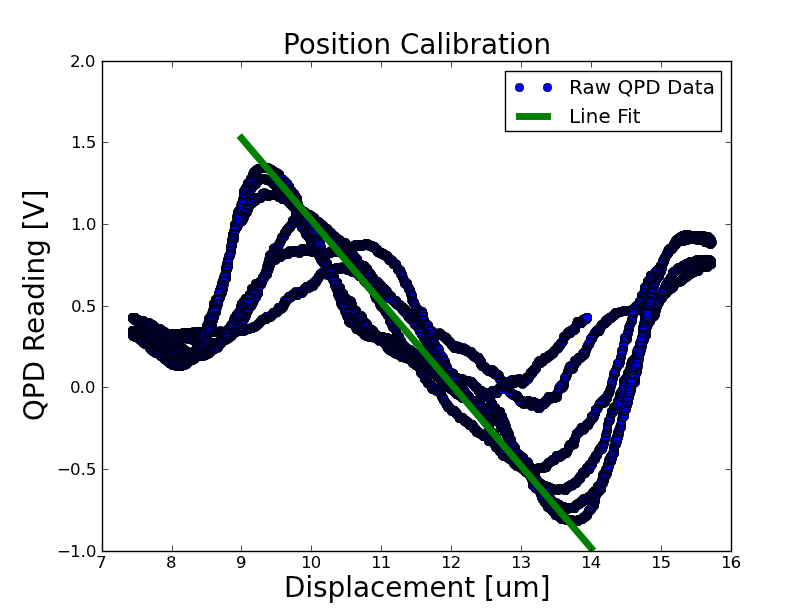
Position Calibration
- Learned to make microfluidic devices out of double-stick tape and a slide -- this was really neat!
- Made two samples with 1 um microspheres
- One sample with free-floating spheres (in H2O)
- Another one with 'stuck' spheres in NaCl (I cheated :( )
- Took a position calibration on the optical trap setup, seen below
calibration: 502463 Volts / m = 1.99 um / volt
Trap Stiffness Calibration
- Did all three versions of the trap calibration, resulting in the following trap spring constants:
| Method | Trap Stiffness (pN / nm) |
|---|---|
| Stokes | 2.26e-5 pN/nm |
| Equipartition Theorem | 1.23e-5 pN/nm |
| PSD | *Need Bandwidth of DAQ* |
Experiments
E Coli
- Learned to culture E Coli (sort of)
- Examined the various cultures to determine which ones had the fastest spinners / spinners in the appropriate direction with Steve
- E Coli weren't spinning very well -- swapped out the blue LED for a Red LED to determine if the wavelength made a difference
- As far as we could tell, the LED color made no difference
- We also couldn't figure out why the E Coli were spinning so slowly
- Worked with Steve to cut the flagella of E Coli by drawing them in and out of a pipette multiple times
- This improved the spinning frequency of the E Coli!
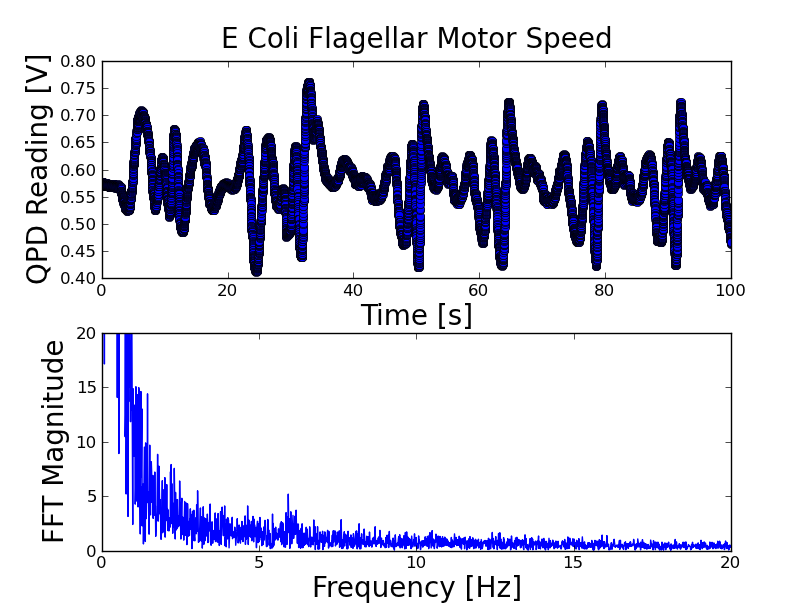
The frequencies here are not properly calibrated until I get the DAQ bandwidth. There are obvious peaks at what are (now) 10, 20, and 40 Hz -- I think these are the harmonics of the e coli rotation, and once the abscissa is properly scaled will give the e coli rotation speed. There looks like there is also a peak hidden in the DC peak.
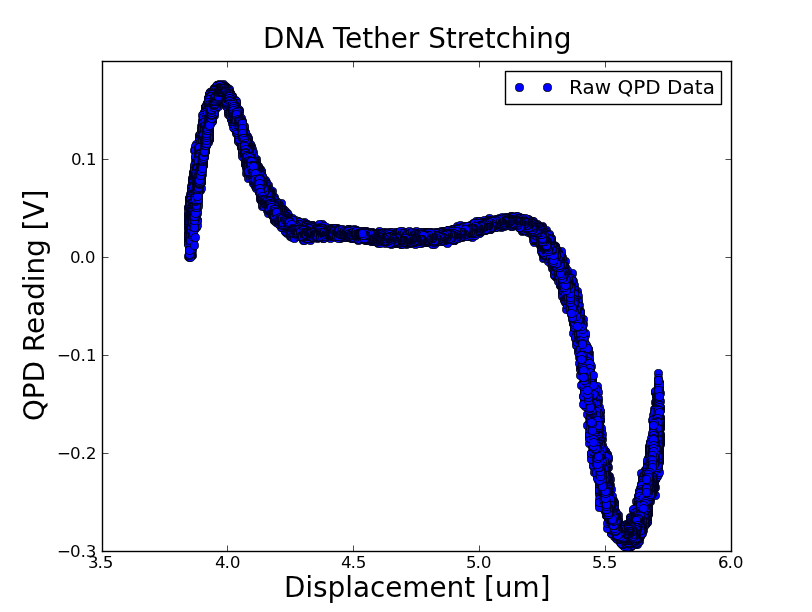
DNA Tethers
- Learned to make the DNA tethers
- Was able to trap a microsphere tethered to the slide via DNA
Stretchiness of the DNA: