Difference between revisions of "DNA Melting: Model function and parameter estimation by nonlinear regression"
| Line 44: | Line 44: | ||
<center><math>S(t)=1-\bar{S}(t)=1-K_{bleaching}\int_0^t f(t) \mathrm{d}t</math></center> | <center><math>S(t)=1-\bar{S}(t)=1-K_{bleaching}\int_0^t f(t) \mathrm{d}t</math></center> | ||
| − | |||
==Evaluating the model== | ==Evaluating the model== | ||
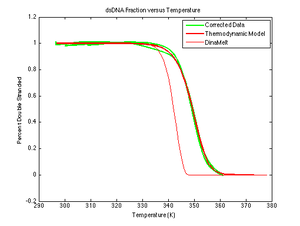
[[Image:Corrected DNA data.png|thumb|right|300 px]] | [[Image:Corrected DNA data.png|thumb|right|300 px]] | ||
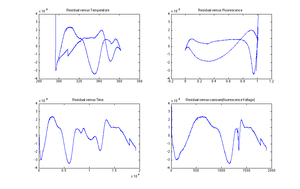
[[Image:Residual plot for DNA data.png|thumb|right|300 px]] | [[Image:Residual plot for DNA data.png|thumb|right|300 px]] | ||
| + | {{Template:20.309 bottom}} | ||
Revision as of 14:12, 16 November 2012
This subject matter will be explained in lecture, in tutorials, and in the lab.
Overview
In addition to the dsDNA concentration, the fluorescence voltage output, $ V_f $, depends of several factors, including:
- dynamics of the temperature cycling system
- $ \Delta H^{\circ} $ and $ \Delta S^{\circ} $
- thermal quenching of the fluorophore
- photobleaching
- responsivity and offset of the instrument
The goal is to write a model for $ V_f $ that takes these effects into account and use nonlinear regression to estimate the parameters.
Photobleaching
Excited fluorescent dye molecules may react chemically with compounds in their environment and permanently lose their ability to fluoresce — a phenomenon called photobleaching. LED and ambient light illuminate the SYBR Green dye for 15 or more minutes over the course of a single melting and annealing cycle, which results in measurable photobleaching. Bleaching can be diminished by lowering the illumination; however, the signal is also proportionally reduced. Photobleaching can also be corrected by a mathematical model.
Assumptions
Photobleaching is a complicated phenomenon. The model proposed here adheres to Dr. George E. P. Box's excellent advice on modeling. Some of the assumptions are more suspect than others.
Which assumptions are probably significant sources of systematic error?
- The dye is divided into two populations, bleached and unbleached, with concentrations S and S
- The initial dye concentration is S + S
- Only dye molecules bound to dsDNA may be excited.
- Only excited fluorophore molecules bleach
- An excited fluorophore will either bleach with probability $ p $ or return to the ground state with probability $ 1-p $.
- The constant $ 1-p $ encompasses all of the mechanisms by which the fluorophore returns to the ground state unbleached, including fluorescence, phosphorescence, and non-radiative relaxation.
- Bleaching is irreversible.
- The number of molecules in the excited state is proportional to fluorescence.
- Bleached and unbleached molecules bind to dsDNA with the same affinity.
Model
Under these assumptions, the bleaching rate is proportional to fluorescence.
$ K_{bleaching} $ encompasses several constants, including $ p $, illumination intensity, optical gain of the instrument, and lock-in amplifier gain.
Setting the initial dye concentration to 1, the fraction of unbleached SYBR Green can be calculated by integrating: