Difference between revisions of "Assignment 10, Part 1: Measuring the osmotic shock response of yeast"
Juliesutton (Talk | contribs) (→Add a power saving module to your pinch valve circuit) |
Juliesutton (Talk | contribs) |
||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| + | |||
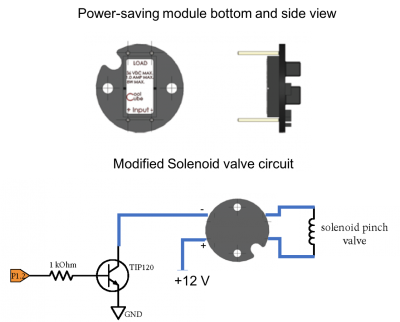
| + | ==Add a power saving module to your pinch valve circuit== | ||
| + | |||
| + | The yeast response to osmotic shock is fairly slow so we'll be switching from low to high salt at very low frequencies (periods up to tens of minutes). You may have noticed in Assignment 8 that the pinch valves get quite hot if they are engaged for a long time. This is a problem because when the tubing gets hot, air bubbles can form in the medium. We want to avoid air bubbles at all costs. | ||
| + | |||
| + | To engage the solenoid valve, we need power delivered from a +12V power supply. Once the solenoid is engaged, however, it requires much less power to remain engaged. The power module shown in the figure below is designed to supply a +12V signal for 100ms, then it drops the power delivered to the solenoid by about 1/3. In this state, the valve will still stay open, but it will produce much less heat (and fewer bubbles!). | ||
| + | |||
| + | [[Image:PinchValvePowerModule.png|center|thumb|400px| reducing power consumption when controlling the solenoid valve ]] | ||
| + | # Modify your pinch valve circuit to use the power module as shown in the figure. Be careful to connect the +12V to the appropriate pin! | ||
| + | # Test that your computer controls still work for the pinch valve and LED illuminators | ||
==Assemble a microfluidic device== | ==Assemble a microfluidic device== | ||
| Line 7: | Line 17: | ||
We'll reuse the tubing you cut in Assignment 8, with some slight modifications. We want to be able to wash the flow channel with ethanol, water and ConA solutions, so we'll connect some short sections of tubing to the device before hooking it up to the media reservoirs. | We'll reuse the tubing you cut in Assignment 8, with some slight modifications. We want to be able to wash the flow channel with ethanol, water and ConA solutions, so we'll connect some short sections of tubing to the device before hooking it up to the media reservoirs. | ||
# For each inlet tubing saved from Assignment 8, cut the shortest section of tygon tubing roughly in half. | # For each inlet tubing saved from Assignment 8, cut the shortest section of tygon tubing roughly in half. | ||
| − | # Connect a second ~1 cm length of PEEK tubing to newly cut short sections of tygon tubing, then insert these | + | # Connect a second ~1 cm length of PEEK tubing to the newly cut short sections of tygon tubing, then insert these into the inlet holes of the PDMS. |
# Similarly, cut the outlet tubing to be roughly 1.5 - 2" long, and insert the orange PEEK tubing into the outlet hole. | # Similarly, cut the outlet tubing to be roughly 1.5 - 2" long, and insert the orange PEEK tubing into the outlet hole. | ||
| Line 15: | Line 25: | ||
# Thaw 1 aliquot of ConA (250 uL of 1 mg/ml in PBS), preferably slowly, on ice. | # Thaw 1 aliquot of ConA (250 uL of 1 mg/ml in PBS), preferably slowly, on ice. | ||
| − | # | + | # Wash your flow channel with 70% ehtanol |
| − | #* Push the | + | #* Load a 1 ml syringe with about 500 uL of 70% ethanol. |
| + | #* Push the ethanol backwards through the device, from the outlet to the inlet, holding the inlets over a wast container. | ||
# Next, wash with ~500 uL DI water. | # Next, wash with ~500 uL DI water. | ||
| − | # Load the ConA solution into a 1 ml syringe. Without introducing any bubbles, | + | # Load the ConA solution into a 1 ml syringe. Without introducing any bubbles, push the entire aliquot of ConA through the device. Leave the syringe connected an let incubate for at least 20 minutes (or until ready to load the yeast cells). |
| − | + | ==Set up media reservoirs and prime inlet tubing== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | ==Set up media reservoirs and prime inlet | + | |
It's common in fluidics experiments to "prime the lines" which means to fill up the tubing with liquid before connecting your microfluidic device. This step can help to prevent bubbles from getting lodged in your device. We want to avoid bubbles at all costs! | It's common in fluidics experiments to "prime the lines" which means to fill up the tubing with liquid before connecting your microfluidic device. This step can help to prevent bubbles from getting lodged in your device. We want to avoid bubbles at all costs! | ||
| Line 45: | Line 46: | ||
The final setup steps include connecting the fluidic device to the reservoirs, then loading the yeas cells into the device. Care should be taken to avoid introducing bubbles, or applying too much pressure once the yeast cells are immobilized. | The final setup steps include connecting the fluidic device to the reservoirs, then loading the yeas cells into the device. Care should be taken to avoid introducing bubbles, or applying too much pressure once the yeast cells are immobilized. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Connect device to fluid reservoirs=== | === Connect device to fluid reservoirs=== | ||
# Attach the high salt reservoir tubing to one inlet tube of the PDMS device. | # Attach the high salt reservoir tubing to one inlet tube of the PDMS device. | ||
| Line 59: | Line 53: | ||
# Engage the valve to open the high salt reservoir and pull some of the high salt medium through the device. | # Engage the valve to open the high salt reservoir and pull some of the high salt medium through the device. | ||
# Toggle the valves several times and try to remove any visible bubbles by (gently!) pulling on the syringe. | # Toggle the valves several times and try to remove any visible bubbles by (gently!) pulling on the syringe. | ||
| + | # Leave the valve in the disengaged (low salt) state, and keep the syringe connected while you prepare the cells. | ||
| + | === Prepare yeast cells=== | ||
| + | |||
| + | # Remove 2x 0.75 mL of the cell culture into two microcentrifuge tubes. | ||
| + | #* The ideal yeast concentration for log-growth is for it to have an OD600 between 0.4 and 0.6. | ||
| + | # Add 0.75 ml of YPD to each culture tube. (The YPD medium helps the cells to form a better pellet.) | ||
| + | # Put both tubes in the microcentrifuge (so that they're balanced) and spin at 800g for 2 minutes. | ||
| + | # Aspirate the supernatant and resuspend one of the pellets in 125 uL of SC medium. Combine the suspended culture with the second pellet, and resuspend it in the same 125uL solution. | ||
| + | |||
=== Load the yeast cells into fluidic device=== | === Load the yeast cells into fluidic device=== | ||
| − | # | + | # Double check that the tubing and device are bubble free, then load the yeast cells into a syringe. |
# Ensure the pinch valve is disengaged (low salt) and slowly load the yeast cells into the device through the outlet tubing, trying not to introduce any bubbles. | # Ensure the pinch valve is disengaged (low salt) and slowly load the yeast cells into the device through the outlet tubing, trying not to introduce any bubbles. | ||
| − | # Leave the syringe connected, and let the cells settle and adhere to the coverslip for 10-15 minutes | + | # Leave the syringe connected, and let the cells settle and adhere to the coverslip for 10-15 minutes. (You can jump ahead to ''Find the focus and set the imaging parameters'' if you want to while you wait.) |
| − | # When ready, disconnect the syringe and connect a long section of tygon tubing to the outlet and | + | # When ready, disconnect the syringe and connect a long section of tygon tubing to the outlet and put the other end in the waste reservoir. |
# You should see excess yeast cells flowing through your device once the outlet is connected. You may need to raise the inlet reservoirs to initiate flow. | # You should see excess yeast cells flowing through your device once the outlet is connected. You may need to raise the inlet reservoirs to initiate flow. | ||
| − | # Toggle the valve several times to ensure that both media reservoirs are flowing as expected. | + | # Toggle the valve several times to ensure that both media reservoirs are flowing as expected, but make sure to leave the cells to equilibrate at low salt. |
==Record movies== | ==Record movies== | ||
| Line 75: | Line 78: | ||
===Find the focus and set the imaging parameters=== | ===Find the focus and set the imaging parameters=== | ||
# Check the alignment of both blue and green illumination for your microscope using a reference slide. | # Check the alignment of both blue and green illumination for your microscope using a reference slide. | ||
| − | # Clip the flow channel onto the stage (use a red acrylic ring as support), and use the room lights (or high gain setting) to | + | # Clip the flow channel onto the stage (use a red acrylic ring as support), and use the room lights (or high gain setting) to bring the yeast cells into focus. |
| − | # Cover your flow channel with a box to block out background light. | + | # Cover your flow channel with a box to block out background light (careful that the box doesn't pull on the tubing). |
# Turn the blue light on, and using a high gain setting, focus on the cells. | # Turn the blue light on, and using a high gain setting, focus on the cells. | ||
| − | # Turn on the green excitation, and again using high gain, adjust the focus. The cell nuclei are | + | # Turn on the green excitation, and again using high gain, adjust the focus. The cell nuclei are harder to focus on, so make sure to always use the green illumination as your final focusing stage. If you don't see a good nucleus image, there's no hope for collecting good data. |
| − | # Lock the stage in all three directions using the locking screws. This is extremely important for long experiment times to prevent the stage from drifting out of focus. | + | # Lock the stage in all three directions using the locking screws. This is ''extremely important'' for long experiment times to prevent the stage from drifting out of focus. |
| − | # With green illumination use the <tt>SetGreenImageParameters</tt> command to set the exposure time to 5s (5000000 us) and adjust the gain to get the best image. | + | # With the green illumination on, use the <tt>SetGreenImageParameters</tt> command to set the exposure time to 5s (5000000 us) and adjust the gain to get the best image. |
# Turn on the blue illuminator and set the exposure and gain. In both cases, exposure times longer than 5 or 6 start to become impractical, so we can only increase gain at that point. | # Turn on the blue illuminator and set the exposure and gain. In both cases, exposure times longer than 5 or 6 start to become impractical, so we can only increase gain at that point. | ||
| − | ===Start | + | ===Start recording data!=== |
| + | |||
| + | Each group will be assigned three conditions to measure. The options are: | ||
| + | # T = 2 min. Record 5 periods, and acquire 4 images per period. | ||
| + | # T = 4 min. Record 5 periods, and acquire 8 images per period. | ||
| + | # T = 8 min. Record 3 periods, and acquire 8 images per period. | ||
| + | # T = 16 min. Record 2 periods, and acquire 8 images per period. | ||
| + | # Step response. Record for 2 minutes at low salt and 20 minutes at high salt. Acquire an image every 2 min. | ||
| + | |||
| + | ''Tips'': | ||
| + | * Start with the smallest period and work your way up. | ||
| + | * Re-check your focus and camera settings after each measurement. You may also want to move your FOV to image fresh cells if the current ones seem dim. | ||
| + | * Don't forget to save your data to the workspace and SAVE YOUR WORKSPACE FREQUENTLY! | ||
| + | * Lock your stage. | ||
==Analyze data== | ==Analyze data== | ||
| + | |||
| + | # For each oscillation movie: | ||
| + | #* Use your code from Assignment 9 to extract the Hog1-response vs. time. | ||
| + | #* On one set of axes, plot the Hog1-response vs. time and the best fit sinusoid. | ||
| + | #* Report the best-fit amplitude, phase, and offset. | ||
| + | # Pool your amplitude and response data with other groups, and make a Bode Plot (amplitude and phase) of the Hog1-response as a function of frequency. | ||
Revision as of 15:48, 19 November 2018
Add a power saving module to your pinch valve circuit
The yeast response to osmotic shock is fairly slow so we'll be switching from low to high salt at very low frequencies (periods up to tens of minutes). You may have noticed in Assignment 8 that the pinch valves get quite hot if they are engaged for a long time. This is a problem because when the tubing gets hot, air bubbles can form in the medium. We want to avoid air bubbles at all costs.
To engage the solenoid valve, we need power delivered from a +12V power supply. Once the solenoid is engaged, however, it requires much less power to remain engaged. The power module shown in the figure below is designed to supply a +12V signal for 100ms, then it drops the power delivered to the solenoid by about 1/3. In this state, the valve will still stay open, but it will produce much less heat (and fewer bubbles!).
- Modify your pinch valve circuit to use the power module as shown in the figure. Be careful to connect the +12V to the appropriate pin!
- Test that your computer controls still work for the pinch valve and LED illuminators
Assemble a microfluidic device
Follow the instructions in Assignment 8 to assemble a PDMS/tape device. Note that you should still have your slab of cured PDMS left over from Assignment 8, so you don't have to repeat that step.
We'll reuse the tubing you cut in Assignment 8, with some slight modifications. We want to be able to wash the flow channel with ethanol, water and ConA solutions, so we'll connect some short sections of tubing to the device before hooking it up to the media reservoirs.
- For each inlet tubing saved from Assignment 8, cut the shortest section of tygon tubing roughly in half.
- Connect a second ~1 cm length of PEEK tubing to the newly cut short sections of tygon tubing, then insert these into the inlet holes of the PDMS.
- Similarly, cut the outlet tubing to be roughly 1.5 - 2" long, and insert the orange PEEK tubing into the outlet hole.
Incubate device with ConA
We need to immobilize yeast cells to the coverslip of the flow device in order to image them over long times. A standard method of immobilizing yeast cells for microscopy is to coat the coverslip with Concanavalin A (ConA) - a lectin that binds to cell surface saccharides.
- Thaw 1 aliquot of ConA (250 uL of 1 mg/ml in PBS), preferably slowly, on ice.
- Wash your flow channel with 70% ehtanol
- Load a 1 ml syringe with about 500 uL of 70% ethanol.
- Push the ethanol backwards through the device, from the outlet to the inlet, holding the inlets over a wast container.
- Next, wash with ~500 uL DI water.
- Load the ConA solution into a 1 ml syringe. Without introducing any bubbles, push the entire aliquot of ConA through the device. Leave the syringe connected an let incubate for at least 20 minutes (or until ready to load the yeast cells).
Set up media reservoirs and prime inlet tubing
It's common in fluidics experiments to "prime the lines" which means to fill up the tubing with liquid before connecting your microfluidic device. This step can help to prevent bubbles from getting lodged in your device. We want to avoid bubbles at all costs!
- Insert the high and low salt media reservoirs into the 50 ml conical tube holders.
- Feed the long length of inlet tubing (tygon-silicone-tygon) into the high-salt reservoir. Make sure the end of the tube is at the bottom of the reservoir. Consider using lab tape to prevent it from getting dislodged.
- Connect a 1 ml syringe to the opposite end of the tubing using a 23G blunt-tipped needle, and pull the fluid from the reservoir into the tubing. Leave the syringe connected.
- Engage the solenoid valve (i.e. OpenHighSalt), then feed the flexible silicone section of tubing into the now-open slot of the pinch valve.
- Close the pinch valve (OpenLowSalt) to prevent flow from the high-salt reservoir, then disconnect the needle and syringe.
- Repeat steps 2-3 with the low-salt reservoir. Feed the silicone section of tubing into the low salt slot of the pinch valve. Leave the syringe connected until ready to connect the PDMS device.
Load yeast cells into the flow channel
The final setup steps include connecting the fluidic device to the reservoirs, then loading the yeas cells into the device. Care should be taken to avoid introducing bubbles, or applying too much pressure once the yeast cells are immobilized.
Connect device to fluid reservoirs
- Attach the high salt reservoir tubing to one inlet tube of the PDMS device.
- Engage the pinch valve to open the high salt valve and close the low salt valve.
- Disconnect the syringe from the low salt tubing and connect it to the remaining inlet tubing of the device.
- Connect the syringe to the outlet of the device, and (gently!) pull some fluid from the low salt reservoir through the device.
- Engage the valve to open the high salt reservoir and pull some of the high salt medium through the device.
- Toggle the valves several times and try to remove any visible bubbles by (gently!) pulling on the syringe.
- Leave the valve in the disengaged (low salt) state, and keep the syringe connected while you prepare the cells.
Prepare yeast cells
- Remove 2x 0.75 mL of the cell culture into two microcentrifuge tubes.
- The ideal yeast concentration for log-growth is for it to have an OD600 between 0.4 and 0.6.
- Add 0.75 ml of YPD to each culture tube. (The YPD medium helps the cells to form a better pellet.)
- Put both tubes in the microcentrifuge (so that they're balanced) and spin at 800g for 2 minutes.
- Aspirate the supernatant and resuspend one of the pellets in 125 uL of SC medium. Combine the suspended culture with the second pellet, and resuspend it in the same 125uL solution.
Load the yeast cells into fluidic device
- Double check that the tubing and device are bubble free, then load the yeast cells into a syringe.
- Ensure the pinch valve is disengaged (low salt) and slowly load the yeast cells into the device through the outlet tubing, trying not to introduce any bubbles.
- Leave the syringe connected, and let the cells settle and adhere to the coverslip for 10-15 minutes. (You can jump ahead to Find the focus and set the imaging parameters if you want to while you wait.)
- When ready, disconnect the syringe and connect a long section of tygon tubing to the outlet and put the other end in the waste reservoir.
- You should see excess yeast cells flowing through your device once the outlet is connected. You may need to raise the inlet reservoirs to initiate flow.
- Toggle the valve several times to ensure that both media reservoirs are flowing as expected, but make sure to leave the cells to equilibrate at low salt.
Record movies
Once you're sure the flow is behaving as expected, let the yeast flow under low salt conditions while you align your microscope and prepare to record data.
Find the focus and set the imaging parameters
- Check the alignment of both blue and green illumination for your microscope using a reference slide.
- Clip the flow channel onto the stage (use a red acrylic ring as support), and use the room lights (or high gain setting) to bring the yeast cells into focus.
- Cover your flow channel with a box to block out background light (careful that the box doesn't pull on the tubing).
- Turn the blue light on, and using a high gain setting, focus on the cells.
- Turn on the green excitation, and again using high gain, adjust the focus. The cell nuclei are harder to focus on, so make sure to always use the green illumination as your final focusing stage. If you don't see a good nucleus image, there's no hope for collecting good data.
- Lock the stage in all three directions using the locking screws. This is extremely important for long experiment times to prevent the stage from drifting out of focus.
- With the green illumination on, use the SetGreenImageParameters command to set the exposure time to 5s (5000000 us) and adjust the gain to get the best image.
- Turn on the blue illuminator and set the exposure and gain. In both cases, exposure times longer than 5 or 6 start to become impractical, so we can only increase gain at that point.
Start recording data!
Each group will be assigned three conditions to measure. The options are:
- T = 2 min. Record 5 periods, and acquire 4 images per period.
- T = 4 min. Record 5 periods, and acquire 8 images per period.
- T = 8 min. Record 3 periods, and acquire 8 images per period.
- T = 16 min. Record 2 periods, and acquire 8 images per period.
- Step response. Record for 2 minutes at low salt and 20 minutes at high salt. Acquire an image every 2 min.
Tips:
- Start with the smallest period and work your way up.
- Re-check your focus and camera settings after each measurement. You may also want to move your FOV to image fresh cells if the current ones seem dim.
- Don't forget to save your data to the workspace and SAVE YOUR WORKSPACE FREQUENTLY!
- Lock your stage.
Analyze data
- For each oscillation movie:
- Use your code from Assignment 9 to extract the Hog1-response vs. time.
- On one set of axes, plot the Hog1-response vs. time and the best fit sinusoid.
- Report the best-fit amplitude, phase, and offset.
- Pool your amplitude and response data with other groups, and make a Bode Plot (amplitude and phase) of the Hog1-response as a function of frequency.