Difference between revisions of "20.109(S24):M1D4"
Noreen Lyell (Talk | contribs) (→Navigation links) |
Noreen Lyell (Talk | contribs) |
||
| Line 67: | Line 67: | ||
#*Record the following information from the spreadsheet: Internal_ID, Zscore_Rep1, and Zscore_Rep2. | #*Record the following information from the spreadsheet: Internal_ID, Zscore_Rep1, and Zscore_Rep2. | ||
| − | ===Part | + | ===Part 4: Choose small molecules for DSF experiment=== |
| + | |||
| + | |||
| + | |||
| + | ====Strategy: Identify common features in small molecules==== | ||
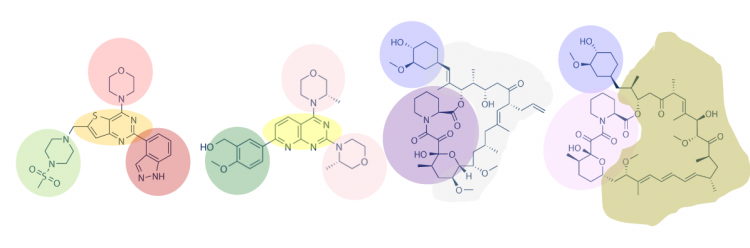
One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. To do this you will carefully examine the hits and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule. | One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. To do this you will carefully examine the hits and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule. | ||
| Line 83: | Line 87: | ||
*How might you make modifications to the small molecules / features to probe binding? As a hint, consider how different functional groups could be positioned at a given site without altering qualitative binding in the SMM assay to translate that into some testable ideas. | *How might you make modifications to the small molecules / features to probe binding? As a hint, consider how different functional groups could be positioned at a given site without altering qualitative binding in the SMM assay to translate that into some testable ideas. | ||
| − | === | + | ====Strategy: Consider the binding metrics from the SMM screen==== |
| + | |||
| + | ====Strategy: Research the biology relevant in Myc:MAX binding==== | ||
| + | |||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(S24):M1D5 |Prepare differential scanning flourimetry (DSF) experiment]] <br> | Next day: [[20.109(S24):M1D5 |Prepare differential scanning flourimetry (DSF) experiment]] <br> | ||
Previous day: [[20.109(S24):M1D3 |Assess purity and concentration of expressed protein]] <br> | Previous day: [[20.109(S24):M1D3 |Assess purity and concentration of expressed protein]] <br> | ||
Revision as of 22:37, 13 February 2024
Contents
Introduction
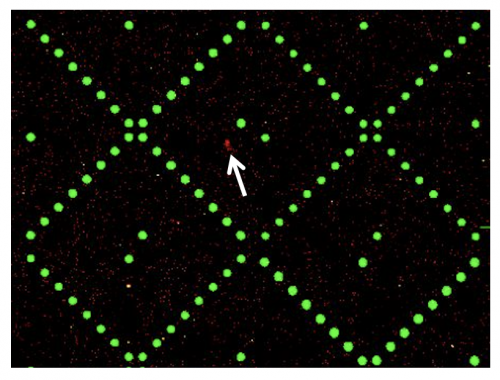
Today you will analyze the data for the SMM screen completed by the Sp23 class of 20.109! For this experiment, students incubated SMM slides with purified MAX-6xHis protein in an effort to identify ligand binders. To identify which of the small molecules, if any, that were screened are able to putatively bind MAX-6xHis, the slides were evaluated using a Genepix microarray scanner. The scanner measured the fluorescence signal emitted from each spot on the slide where a ligand was printed. Considering that neither the ligands nor MAX protein is fluorescent, from where does this signal arise?
The above image is from raw data generated during the SMM screen. The green spots represent locations on the SMM slide where fluorescein was printed. As a reminder, fluorescein is a fluorescent dye used for alignment purposes. Correct alignment is critical to knowing which ligands are in which spots on the slide. Red spots are indicative of ligands that are bound by MAX-6xHis (example denoted by white arrow in above image). The signal is due to the Alexa Fluor® 647, a fluorophore that was conjugated to an antibody that is specific to the 6xHis tag.
When the slides are imaged, the scanner exposes the SMM to excitation light specific to the fluorophores used in the experiment (i.e. wavelengths that excite fluorescein and the Alexa Fluor 647 bound to MAX-6xHis). The scanner then detects the intensity of the emitted fluorescence light to generate an image that can be analyzed. This analysis will provide a list of 'hits' that can be used to direct future studies.
Protocols
Part 1: Participate in Communication Lab workshop
Our communication instructor, Dr. Chiara Ricci-Tam, will join us today to focus on figure design.
Part 2: Analyze results of SMM screens
After the Sp23 class completed the SMM slide preparation steps, the Instructors imaged the slides and completed the initial data analysis as outlined in prelab. Here you will complete the data analysis to review the hits identified for MAX-6xHis.
- Download the SMM results .csv file from the class data page, and save it to the desktop with the jupyter notebook file.
- Open Anaconda Navigator.
- On the home screen, you will first change the environment.
- In the dropdown that shows 'base (root)', select 'SMM_Analysis'.
- Launch the Jupyter notebook by clicking "launch" in the appropriate panel.
- Do not interrupt the terminal!
- A web browser will open when the the Jupyter notebook is active.
- Select Desktop folder.
- Select Sp23 SMM Analysis.ipynb.
- Before running the Jupyter notebook analysis pipeline, you will need to enter the name of the appropriate .csv file.
- Go to In [5] and delete 'HECT_screen.csv'.
- Enter 'screen_results.csv'.
- To run the analysis, select In [1] of the code and click 'Run' from the toolbar at the top of the Jupyter notebook window.
- Wait for the analysis to complete before running the next section.
- When the analysis from In [1] is running, an asterisk will appear in the place of the 1. When the analysis is completed the 1 will reappear.
- Read through the content provided after In [1].
- Then complete each step of the analysis (one at a time!) by completing the Step #8 for each of the cells.
- Be sure the content provided for each step of the analysis as you go.
- When you reach the section titled "Histograms - Finding the Z score threshold" you will use the preset threshold of 10 for the data analysis.
- After you run this analysis two histograms will appear in the cell.
- Using the information provided in prelab you will refine the threshold to better define hits attained in your experiment.
- To repeat the analysis with your refined threshold, enter the value you decide after 'zscore_h = ' in the cell.
- The default value of 10 will be present, simply delete and enter your value.
- Rerun the selected cell.
- In your laboratory notebook, complete the following:
- Record the threshold value selected.
- Attach screen captures of the histograms.
- The run from the section titled "Scatter Plot - Data distribution and consistency between replicates" will generate a scatter plot.
- In your laboratory notebook, complete the following:
- Provide an assessment of the distribution based on the prelab discussion.
- The run from the section titled "Hits: calling, visualizing and validating" will generate a table with the information for the hits to MAX-6xHis.
- In the 'Calling hits' sub-section, the analysis will result in a hit rate for your screen. Read the content provided below the hit rate to better refine your list of hits according to the ideal hit rate.
- In your laboratory notebook, complete the following:
- Record initial hit rate from the default threshold value.
- Describe the changes made to the threshold based on the initial rate and record the final threshold value you chose.
- Record the final hit rate determined after changing the threshold.
- In the 'Visualize hits structures with rdkit' sub-section, the analysis will show chemical structures for the hits.
- To complete visual identification of common features, you can download the image to see all of the structures together.
- You will use this for Part 2!
- In the 'Validating hits' sub-section, the analysis will provide a table with information for each of the hits.
- The by-eye analysis described was completed for you as specific software is needed to visualize the individual spots. Read through the direction so you know how this was done.
- In your laboratory notebook, complete the following:
- Record the following information from the spreadsheet: Internal_ID, Zscore_Rep1, and Zscore_Rep2.
Part 4: Choose small molecules for DSF experiment
Strategy: Identify common features in small molecules
One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. To do this you will carefully examine the hits and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule.
Review the hits that were identified in the SMM screen completed for MAX-6xHis. Use the Internal_ID number from the spreadsheet to find the chemical structures of the hits in the image saved in Part #1, Step #19.
In your laboratory notebook, complete the following:
- How many features did you identify that are present in two or more of the small molecules that putatively bind MAX-6xHis? Are there more or less than you expected?
- Is there a feature present in all of the identified small molecules? What might this suggest about the binding site(s) and / or binding ability of MAX?
- Can you assign the identified small molecules to sub-groups based on the common features that are present?
- What might the different features represent? More specifically, consider whether each subgroup has a unique binding site on the target protein or if each subgroup represents different solutions for interacting with the same binding site.
- How might you make modifications to the small molecules / features to probe binding? As a hint, consider how different functional groups could be positioned at a given site without altering qualitative binding in the SMM assay to translate that into some testable ideas.
Strategy: Consider the binding metrics from the SMM screen
Strategy: Research the biology relevant in Myc:MAX binding
Next day: Prepare differential scanning flourimetry (DSF) experiment