Difference between revisions of "20.109(S23):M2D4"
Becky Meyer (Talk | contribs) (→Part 1: Examine psgRNA_target sequencing results) |
Becky Meyer (Talk | contribs) (→Protocols) |
||
| Line 51: | Line 51: | ||
*Attach a screenshot for each alignment. | *Attach a screenshot for each alignment. | ||
*Record which clone contains the correct point mutation. | *Record which clone contains the correct point mutation. | ||
| + | |||
| + | |||
| + | ===Part 2: Prepare coverslips for immunofluorescence staining=== | ||
| + | |||
| + | While the yeast transformations are growing, you will prepare materials to confirm transporter expression in our yeast model system. As preparation for the IF experiment on M2D6, you will work in a sterile biosafety hood to coat coverslips for immunofluorescence staining. | ||
| + | |||
| + | #Collect a 12 well plate, a set of coverslips, and fine-tip forceps. | ||
| + | #*These materials are packaged to maintain sterility and should only be opened in the hood. | ||
| + | #*Remember to spray down all materials before placing them in the hood! | ||
| + | #Using the forceps, add coverslips to the 12 well plate according to the chart. | ||
| + | #Take an aliquot of sterile poly-D-lysine (PDL) into the hood. | ||
| + | #Add 1ml of PDL to each well that contains a coverslip. | ||
| + | #Allow coverslips to incubate with PDL for 20 minutes in the hood. | ||
| + | #Following incubation, aspirate the PDL and wash coverslips 2x with 1ml of DI water. | ||
| + | #Aspirate the final wash and allow the coverslips to dry in the hood for 20 minutes. | ||
| + | #Once coated coverslips have dried, label the plate with your team name/section and wrap the plate in parafilm for storage. | ||
| + | #Store plates at 4°C. | ||
| + | |||
| + | |||
| + | ===Part 3: Prepare reagents for metal incubation=== | ||
| + | |||
| + | ==Reagents list== | ||
| + | *Poly-D-Lysine 0.1mg/ml (Gibco) | ||
| + | **Synthetic dropout-uracil (SD-U) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.192 % amino acid mix lacking uracil (Sigma), 2% glucose (BD Bacto), 0.1% adenine hemisulfate (Sigma) | ||
| + | **Cadmium chloride (Sigma), [stock] = 100mM | ||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(S23):M2D5 |Analyze ICP-OES data]] <br> | Next day: [[20.109(S23):M2D5 |Analyze ICP-OES data]] <br> | ||
Previous day: [[20.109(S23):M2D3 |Sequence clones and transform into yeast cells]] <br> | Previous day: [[20.109(S23):M2D3 |Sequence clones and transform into yeast cells]] <br> | ||
Revision as of 18:34, 2 February 2023
Contents
Introduction
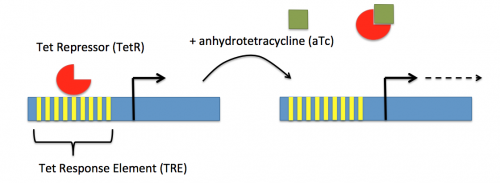
In contrast, expression of the gene encoding dCas9 within pdCas9 is regulated by an inducible promoter (pLtetO-1). An inducible promoter is 'off' unless the appropriate molecule is present to relieve repression. In the case of the CRISPRi system, expression of the gene encoding dCas9 is inhibited due to the use of a tet-based promoter construct. Tet is shorthand for tetracycline, which is an antibiotic that inhibits protein synthesis through preventing the association between charged aminoacyl-tRNA molecules and the A site of ribosomes. Bacterial cells that carry the tet resistance cassette are able to survive exposure to tetracycline by expressing genes that encode an efflux pump that 'flushes' the antibiotic from the bacterial cell. To conserve energy, the tet system is only expressed in the presence of tetracyline. In the absence of tetracycline, a transcription repressor protein (TetR) is bound to the promoter upstream of the tet resistance cassette genes. When tetracycline is present, the molecule binds to TetR causing a confirmational change that results in TetR 'falling off' of the promoter. In the CRISPRi system, the tet-based promoter construct upstream of the gene that encodes dCas9 is 'off' unless anhydrotetracyline (aTc), an analog of tetracyline, is added to the culture media. Why is it important to use an analog rather than the actual antibiotic?Taken together, the sgRNA_target is constitutively transcribed and, thereby, always present. The dCas9 protein is only present when aTc is added. Thus, gene expression is only altered when aTc is present. In this, when dCas9 is expressed it forms a complex with the sgRNA_target. The sgRNA target then 'seeks out' the target within the host genome. When the targeted sequence is recognized, the complex binds and acts as a 'roadblock' by prohibiting RNAP access to the sequence. Because the targeted gene is not able to be transcribed, the protein encoded by that gene is not synthesized. In our experiments, we hypothesize that the absence of specific proteins, or enzymes, involved in anaerobic fermentative metabolism will increase the yield of ethanol.
Protocols
Part 1: Examine Fet4_mutant sequencing results
Your goal today is to analyze the sequencing data for you two potential mutant Fet4 clones - two independent colonies from your cloning reaction - and then decide which colony to proceed with to engineer yeast to become a cadmium sink.
Retrieve Fet4_mutant sequence results from Genewiz
- Your sequencing data is available from Genewiz. For easier access, the information was uploaded to the [20.109(S22):Class_data Class Data tab].
- Download the zip folder with your team sequencing results and confirm that there are 8 files saved in the folder.
- For each sequencing reaction, you should have one .ab1 file and one .seq file.
- Open one of the .ab1 files.
- This file contains the chromatogram for your sequencing reaction. Scroll through the sequence and ensure that the peaks are clearly defined and evenly spaced. Low signal (or peaks) or stacked peaks can provide incorrect base assignments in the sequence.
- Open one of the .seq files.
- This file contains the base assignments for your sequencing reaction. The bases are assigned by the software from the chromatogram sequence.
- The start of the a sequencing reaction result often contains several Ns, which indicates that the software was unable to assign a basepair.
In your laboratory notebook, complete the following:
- Given the chromatogram result, why might the software assign Ns in the start of the sequence?
- Visually inspect the chromatograms for all of your sequencing results.
- Do the peaks appear clearly defined or is there overlap? What might this indicate about the quality of your sequencing results?
- Do the peaks extend above the background signal? What might this indicate about the quality of your sequencing results?
Confirm point mutation in Fet4 sequence using SnapGene
You should align your sequencing data with a known sequence, in this case the Fet4 sequence from M2D1, to identify a successful mutation as well as any unintended base changes that may have occurred. There are several web-based programs for aligning sequences and still more programs that can be purchased. The steps for using SnapGene are below. Please feel free to use any program with which you are familiar.
- Generate a new DNA file that contains the Fet4 sequence you examined on M2D1.
- Generate an additional new DNA file that contains the results from the sequencing reaction completed by Genewiz.
- For each sequencing result you should generate a distinct new DNA file. Remember you should have a forward and reverse sequencing result for each of your clones!
- Paste the sequence text from your sequencing run into the new DNA file window. If there were ambiguous areas of your sequencing results, these will be listed as "N" rather than "A" "T" "G" or "C" and it's fine to include Ns in the query.
- The start and end of your sequencing may have several Ns. In this case it is best to omit these Ns by pasting only the 'good' sequence that is flanked by the ambiguous sequence.
- To confirm the mutation sequence in your clones, open one of the forward sequencing results files generated in the previous step.
- Select 'Tools' --> 'Align to Reference DNA Sequence...' --> 'Align Full Sequences...' from the toolbar.
- In the window, select the file that contains the sgRNA oligo sequence and click 'Open'.
- A new window will open with the alignment of the two sequences. The top line of sequence shows the results of the sequencing reaction and the bottom line shows the oligo you designed.
- Are there any discrepancies or differences between the two sequences? Scroll through the entire alignment to check the full sequencing result and note any basepair changes.
- Follow the above steps to examine all of your sequencing results. Remember: you used a forward and a reverse primer to interrogate both potential Fet4 point mutations.
- From the alignments, determine which Fet_mutant clone has the correct point mutation.
- If both clones contain the correct sequence choose either yeast transformation to use in the rest of your experiments. If only one is correct, then this is the transformant you will use. If neither of your plasmids carry the appropriate mutation, talk to your Instructor.
In your laboratory notebook, complete the following:
- Attach a screenshot for each alignment.
- Record which clone contains the correct point mutation.
Part 2: Prepare coverslips for immunofluorescence staining
While the yeast transformations are growing, you will prepare materials to confirm transporter expression in our yeast model system. As preparation for the IF experiment on M2D6, you will work in a sterile biosafety hood to coat coverslips for immunofluorescence staining.
- Collect a 12 well plate, a set of coverslips, and fine-tip forceps.
- These materials are packaged to maintain sterility and should only be opened in the hood.
- Remember to spray down all materials before placing them in the hood!
- Using the forceps, add coverslips to the 12 well plate according to the chart.
- Take an aliquot of sterile poly-D-lysine (PDL) into the hood.
- Add 1ml of PDL to each well that contains a coverslip.
- Allow coverslips to incubate with PDL for 20 minutes in the hood.
- Following incubation, aspirate the PDL and wash coverslips 2x with 1ml of DI water.
- Aspirate the final wash and allow the coverslips to dry in the hood for 20 minutes.
- Once coated coverslips have dried, label the plate with your team name/section and wrap the plate in parafilm for storage.
- Store plates at 4°C.
Part 3: Prepare reagents for metal incubation
Reagents list
- Poly-D-Lysine 0.1mg/ml (Gibco)
- Synthetic dropout-uracil (SD-U) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.192 % amino acid mix lacking uracil (Sigma), 2% glucose (BD Bacto), 0.1% adenine hemisulfate (Sigma)
- Cadmium chloride (Sigma), [stock] = 100mM
Next day: Analyze ICP-OES data