Difference between revisions of "20.109(S23):M1D7"
Noreen Lyell (Talk | contribs) (→Protocols) |
Noreen Lyell (Talk | contribs) (→Protocols) |
||
| Line 8: | Line 8: | ||
==Protocols== | ==Protocols== | ||
| − | ===Part | + | ===Part 1: Analyze results of SMM screens=== |
| − | + | After you completed the SMM slide preparation steps, the Instructors imaged the slides and completed the initial data analysis as outlined in prelab. Here you will complete the data analysis to identify the hits for MAX-6xHis. | |
| − | + | #Open Anaconda Navigator. | |
| + | #On the home screen, you will first change the environment. | ||
| + | #*In the dropdown that shows 'base (root)', select 'SMM_Analysis'. | ||
| + | #Launch the Jupyter notebook by clicking "launch" in the appropriate panel. | ||
| + | #*Do not interrupt the terminal! | ||
| + | #*A web browser will open when the the Jupyter notebook is active. | ||
| + | #Select Desktop folder. | ||
| + | #Select Sp23 SMM Analysis.ipynb. | ||
| + | #Before running the Jupyter notebook analysis pipeline, you will need to enter the name of the appropriate .csv file. | ||
| + | #*Go to In [5] and delete 'HECT_screen.csv'. | ||
| + | #*Enter 'screen_results.csv'. | ||
| + | #To run the analysis, select In [1] of the code and click 'Run' from the toolbar at the top of the Jupyter notebook window. | ||
| + | #*Wait for the analysis to complete before running the next section. | ||
| + | #*When the analysis from In [1] is running, an asterisk will appear in the place of the 1. When the analysis is completed the 1 will reappear. | ||
| + | #Read through the content provided after In [1]. | ||
| + | #Then complete each step of the analysis (one at a time!) by completing the Steps described for In [1] for each of the cells. | ||
| + | #*Be sure the content provided for each step of the analysis as you go. | ||
| + | #When you reach the section titled "Histograms - Finding the Z score threshold" you will use the preset threshold of 10 for the data analysis. | ||
| + | #*After you run this analysis two histograms will appear in the cell. | ||
| + | #Using the information provided in prelab you will refine the threshold to better define hits attained in your experiment. | ||
| + | #*To repeat the analysis with your refined threshold, enter the value you decide after 'zscore_h = ' in the cell. | ||
| + | #*The default value of 10 will be present, simply delete and enter your value. | ||
| + | #*Rerun the selected cell. | ||
| − | + | # in your notebook. | |
| − | + | #The run from the section titled "Scatter Plot - Data distribution and consistency between replicates" will generate a scatter plot. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | ===Part | + | |
| + | #*Include notes on the distribution based on the prelab discussion in your notebook. | ||
| + | |||
| + | #The run from the section titled "Hits: calling, visualizing and validating" will generate a table with the information for the hits to MAX-6xHis. | ||
| + | #In the 'Calling hits' sub-section, the analysis will result in a hit rate for your screen. Read the content provided below the hit rate to better refine your list of hits according to the ideal hit rate. | ||
| + | |||
| + | # in your notebook record initial hit rate and any modifications to threshold needed to attain ideal hit rate. Record new hit rate after modifications. | ||
| + | |||
| + | #In the 'Visualize hits structures with rdkit' sub-section, the analysis will show chemical structures for the hits. | ||
| + | #*To complete visual identification of common features, you can download the image to see all of the structures together. | ||
| + | #*You will use this for Part 2! | ||
| + | #In the 'Validating hits' sub-section, the analysis will provide a table with information for each of the hits. | ||
| + | #*The by-eye analysis described was completed for you as specific software is needed to visualize the individual spots. Read through the direction so you know how this was done. | ||
| + | |||
| + | ===Part 2: Identify common features in SMM hits=== | ||
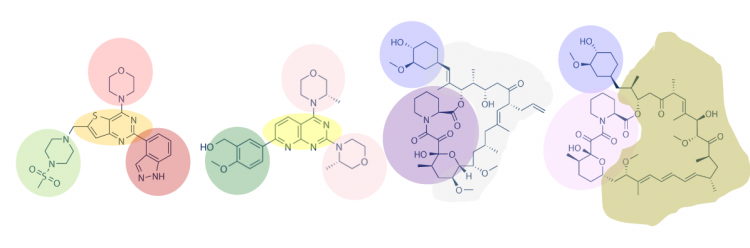
One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. To do this you will carefully examine the hits and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule. | One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. To do this you will carefully examine the hits and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule. | ||
Revision as of 21:12, 6 March 2023
Contents
Introduction
Protocols
Part 1: Analyze results of SMM screens
After you completed the SMM slide preparation steps, the Instructors imaged the slides and completed the initial data analysis as outlined in prelab. Here you will complete the data analysis to identify the hits for MAX-6xHis.
- Open Anaconda Navigator.
- On the home screen, you will first change the environment.
- In the dropdown that shows 'base (root)', select 'SMM_Analysis'.
- Launch the Jupyter notebook by clicking "launch" in the appropriate panel.
- Do not interrupt the terminal!
- A web browser will open when the the Jupyter notebook is active.
- Select Desktop folder.
- Select Sp23 SMM Analysis.ipynb.
- Before running the Jupyter notebook analysis pipeline, you will need to enter the name of the appropriate .csv file.
- Go to In [5] and delete 'HECT_screen.csv'.
- Enter 'screen_results.csv'.
- To run the analysis, select In [1] of the code and click 'Run' from the toolbar at the top of the Jupyter notebook window.
- Wait for the analysis to complete before running the next section.
- When the analysis from In [1] is running, an asterisk will appear in the place of the 1. When the analysis is completed the 1 will reappear.
- Read through the content provided after In [1].
- Then complete each step of the analysis (one at a time!) by completing the Steps described for In [1] for each of the cells.
- Be sure the content provided for each step of the analysis as you go.
- When you reach the section titled "Histograms - Finding the Z score threshold" you will use the preset threshold of 10 for the data analysis.
- After you run this analysis two histograms will appear in the cell.
- Using the information provided in prelab you will refine the threshold to better define hits attained in your experiment.
- To repeat the analysis with your refined threshold, enter the value you decide after 'zscore_h = ' in the cell.
- The default value of 10 will be present, simply delete and enter your value.
- Rerun the selected cell.
- in your notebook.
- The run from the section titled "Scatter Plot - Data distribution and consistency between replicates" will generate a scatter plot.
- Include notes on the distribution based on the prelab discussion in your notebook.
- The run from the section titled "Hits: calling, visualizing and validating" will generate a table with the information for the hits to MAX-6xHis.
- In the 'Calling hits' sub-section, the analysis will result in a hit rate for your screen. Read the content provided below the hit rate to better refine your list of hits according to the ideal hit rate.
- in your notebook record initial hit rate and any modifications to threshold needed to attain ideal hit rate. Record new hit rate after modifications.
- In the 'Visualize hits structures with rdkit' sub-section, the analysis will show chemical structures for the hits.
- To complete visual identification of common features, you can download the image to see all of the structures together.
- You will use this for Part 2!
- In the 'Validating hits' sub-section, the analysis will provide a table with information for each of the hits.
- The by-eye analysis described was completed for you as specific software is needed to visualize the individual spots. Read through the direction so you know how this was done.
Part 2: Identify common features in SMM hits
One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. To do this you will carefully examine the hits and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule.
Review the hits that were identified in the SMM screen completed for TDP43-RRM12. To see the chemical structures, translate the SMILES strings using one of the methods described in the text below the table. It may be easier to copy / paste the small molecule images into a powerpoint file so you can readily see all of the structures. Also, it may be helpful to use a color-coding system (like the one in the image provided above) to highlight features / structures that are common to the hits.
These online resources may be helpful to learning more about the hits identified in the SMM:
- Cloud version of ChemDraw here.
- Copy and paste the small molecule smiles into the work space to get a chemical structure
- Platform to transform the smiles information into a PubChem ID here.
- Copy and paste the smiles into the input ID search to determine the ID number.
- PubChem database of chemical information here.
- Includes small molecule molecular weight and other useful information.
In your laboratory notebook, complete the following:
- How many features did you identify that are present in two or more of the small molecules that putatively bind TDP43-RRM12? Are there more or less than you expected?
- Is there a feature present in all of the identified small molecules? What might this suggest about the binding site(s) and / or binding ability of TDP43-RRM12?
- Can you assign the identified small molecules to sub-groups based on the common features that are present?
- What might the different features represent? More specifically, consider whether each subgroup has a unique binding site on the target protein or if each subgroup represents different solutions for interacting with the same binding site.
- How might you make modifications to the small molecules / features to probe binding? As a hint, consider how different functional groups could be positioned at a given site without altering qualitative binding in the SMM assay to translate that into some testable ideas.
Reagents list
Next day: Organize Data summary figures and results