Difference between revisions of "20.109(S22):M1D8"
Noreen Lyell (Talk | contribs) (→Part 2: Use statistics to interpret results) |
Noreen Lyell (Talk | contribs) (→Part 1: Image and analyze TDP43 localization experiment results) |
||
| Line 62: | Line 62: | ||
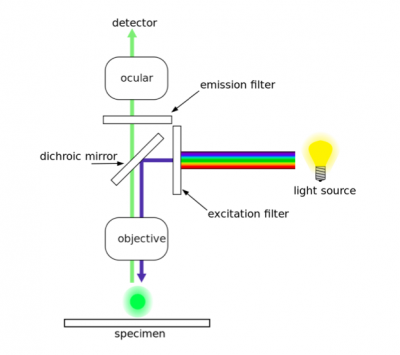
Fluorescence, or epifluorescence, microscopes are composed of a light source, an excitation filter, a dichroic mirror, and an emission filter. The filters and the dichroic mirror are specific to the spectral excitation and emission characteristics of the fluorescent tag. To visualize fluorescence, light at the excitation wavelength is focused on the sample. The light emission from the sample is focused by the objective to a detector. | Fluorescence, or epifluorescence, microscopes are composed of a light source, an excitation filter, a dichroic mirror, and an emission filter. The filters and the dichroic mirror are specific to the spectral excitation and emission characteristics of the fluorescent tag. To visualize fluorescence, light at the excitation wavelength is focused on the sample. The light emission from the sample is focused by the objective to a detector. | ||
| − | <font color = #0d368e>'''To ensure you are familiar with the steps involved in imaging the TDP43-localization experiment, please watch the video tutorial linked here: [[https://www.dropbox.com/s/2akfooztl7ci9qm/H2AX%20Imaging.mp4?dl=0 | + | <font color = #0d368e>'''To ensure you are familiar with the steps involved in imaging the TDP43-localization experiment, please watch the video tutorial linked here: [[https://www.dropbox.com/s/2akfooztl7ci9qm/H2AX%20Imaging.mp4?dl=0 Imaging]].'''</font color> |
For timing reasons, the slides you prepared in the previous laboratory session were imaged prior to class today. The Instructors will provide a demonstration of the imaging process at the microscope so you understand how the data were captured for the analysis steps you will complete below. | For timing reasons, the slides you prepared in the previous laboratory session were imaged prior to class today. The Instructors will provide a demonstration of the imaging process at the microscope so you understand how the data were captured for the analysis steps you will complete below. | ||
Revision as of 13:24, 26 January 2022
Contents
Introduction
Today is the final laboratory session for Module 1! You have completed all of the bench work for your research; however, there is still data analysis to complete for your experiments. In addition to plotting the data, you will complete statistical analysis to determine the significance of your results.
Statistics are mathematical tools used to analyze, interpret, and organize data. The specific tools that you will use are confidence intervals (CI) and the Student's t-test. To begin, review the following definitions:
- Mean (or average) is defined as:
$ \overline{\chi } = \frac{\sum_{i}^{n}\chi _{i}}{n} $, where $ \chi _{i} $ = individual value and n = number of samples
- With infinite data, the mean ($ \overline{\chi } $) approaches the true mean (μ).
- Standard deviation measures the variation in the data and is defined as:
$ s = \sqrt{\frac{\sum_{i}^{n }(\chi _{_{i}}-\overline{\chi })}{n - 1}} $, where n - 1 = degrees of freedom
- With infinite data, the standard deviation (s) approaches the true standard deviation (σ).
Because standard deviation is only justified when sufficient data have been collected to generate a normal curve, you will use confidence intervals to report the likelihood that your results predict the true mean. A confidence interval is a defined interval that is calculated to define the true mean to a specified level of confidence. Simply, it is possible to define a range in your data set that likely contains the true mean based on the calculated mean.
- Confidence interval (CI) is defined as:
CI = $ \overline{\chi } \pm \frac{ts}{\sqrt{n}} $, where t = value from t table (dependent on specified confidence level and n)
In your data, you should use the CI to generate error bars due the low n. Be sure to report which confidence level was used to calculate the intervals reported. So, what does this all mean in regard to the data you will report? As an example, if the calculated $ \overline{\chi } $ of a data set equals 80 au there is a 95% chance the μ is between 50 au and 110 au, where au = arbitrary units. And how does this relate to s? If you know the μ, the σ represents a 68% confidence interval.
When interpreting data, the error bars are representative of the noise in the data or how different the data points are for each of the replicates. Replicates come in two types: technical and biological. Technical replicates indicate that the same sample was tested multiple times and is measure of experimenter error (for example, pipetting errors between aliquots). Biological replicates indicate that different preparations of the same sample were tested and is a measure of the difference in a response to a variable (for example, response to a treatment between separate cultures of the same cell line). Though both types have value in data analysis, the interpretation of the error represented in each case is different. Because of this it is important to indicate if the replicates used in the data analysis are technical or biological. For your data, what type of replicates did you analyze for the γH2AX experiment? For the CometChip experiment?
Lastly, you will use Student's t-test to report if your data are statistically different between treatments.
- Student's t-test is defined as:
$ t = \frac{\left | \overline{\chi_{_{1}}} - \overline{\chi_{_{2}}} \right |}{s_{pooled}}\sqrt{\frac{n_{1} n_{2}}{n_{1}+n_{2}}} $, where $ s_{pooled} = \sqrt{\frac{s_{1}^{2} (n_{1} -1) + {s_{2}^{2} (n_{2} - 1)}{}}{n_{1} + n_{2} - 2}} $
The value you calculate with the Student's t-test equation is referred to as tcalculated. This tcalculated value is compared to the ttabulated value in the the t table, according to the appropriate n - 1 using the p-value for the two-tailed distribution (which assumes that you do not know how the data will shift). If the tcalculated value is greater than the ttabulated, then the data sets are significantly different at the specific p-value. So, what does this all mean in regard to the data you will report? As an example, if the tcalculated for a data set with n - 1 = 10 is 3 (given that the ttabulated is 2.228), then the data sets are different with a p-value ≤ 0.05. Which means that there is less that a 5% chance that the data sets are the same.
Protocols
Part 1: Image and analyze TDP43 localization experiment results
As discussed in prelab, two antibodies were used in the TDP43-localization assay. The first antibody, or primary antibody, was anti-TDP43 and raised in a mouse. The secondary antibody was anti-mouse and raised in a goat, more importantly, this molecule is conjugated to a fluorescent dye tag called Alexa Fluor 488. The Alexa Fluor 488 tag is a bright, green fluorescent dye that is excited at 488 nm. To visualize TDP43 in CAD cells, we will use fluorescence microscopy.In fluorescence microscopy the specimen is illuminated with a wavelength of light specific to the excitation of the fluorescent tag used to target the feature of interest. The excitation wavelength is absorbed by the fluorescent tag, which causes it to emit light at a longer, less energetic wavelength. Typically, fluorescence microscopes used in biology are an epifluorescence type with a single light path (the objective) for excitation and emission detection, as depicted in the diagram above.
Fluorescence, or epifluorescence, microscopes are composed of a light source, an excitation filter, a dichroic mirror, and an emission filter. The filters and the dichroic mirror are specific to the spectral excitation and emission characteristics of the fluorescent tag. To visualize fluorescence, light at the excitation wavelength is focused on the sample. The light emission from the sample is focused by the objective to a detector.
To ensure you are familiar with the steps involved in imaging the TDP43-localization experiment, please watch the video tutorial linked here: [Imaging].
For timing reasons, the slides you prepared in the previous laboratory session were imaged prior to class today. The Instructors will provide a demonstration of the imaging process at the microscope so you understand how the data were captured for the analysis steps you will complete below.
ANALYSIS OF IMAGES STEPS
Part 2: Use statistics to interpret results
Use the statistical analysis tools described in the Introduction to analyze the data for your aggregation and localization experiments. The figures / analyses in your Data summary should include measures of variability (i.e. confidence intervals) and significance (i.e. p-values).
For the aggregation data:
In the aggregation experiment that you completed, you included three replicates for each condition. For the figure that you will include in the Data summary, plot the averaged values then perform statistical analysis to determine the variability and significance of your data.
For the localization data:
XX
In your laboratory notebook, complete the following:
Next day: Complete in-silico cloning of pdCas9 expression plasmid