20.109(S22):M1D1
Contents
Introduction
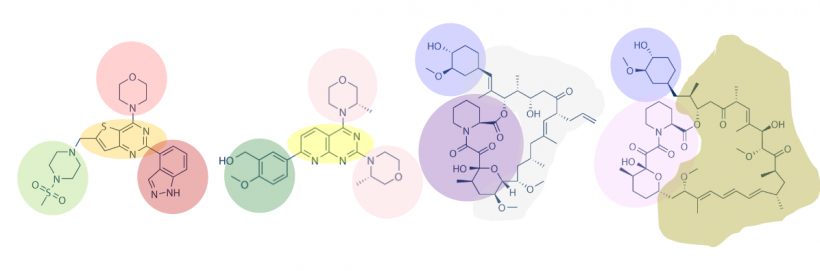
A small molecule microarray (SMM) requires the covalent attachment of a library of small molecules to a glass slide. Our library is meant to broadly sample the drug-like chemical space (i.e. all possible chemical structures that have drug-like physical properties) and contains about 50,000 small molecules. Some libraries are much smaller, while many pharmaceutical companies possess high-throughput screening (HTS) collections of millions of compounds. Because this chemical space is very large, it’s difficult to generalize any single chemical reaction for this attachment that can be applied to all small molecules. We take a “one-size-fits-most” approach, where the glass slide is functionalized with a broadly reactive electrophile capable of reacting with nucleophiles present in most drug-like small molecules, such as alcohols or amines. Many small molecules contain multiple nucleophiles suitable for attachment. In this case, our manufacturing will result in a mixture of attachment sites. It’s important to remember that attachment to the glass slide constrains the possible orientation of the protein-small molecule interaction; some orientations are not possible because the glass slide and linker are in the way.
We start with a glass slide with exposed amines across the surface and attach a short PEG (polyethylene glycol) linker. To the end of this PEG linker, an isocyanate group is attached. Isocyanate, or R-N=C=O, is a resonant structure, and a partial positive charge is stabilized on the carbon atom. This carbon atom is electrophilic, and small molecules with nucleophiles will react here. We estimate that about 70% of drug-like small molecules are amenable to this reaction, and our library is filtered to contain only these molecules.
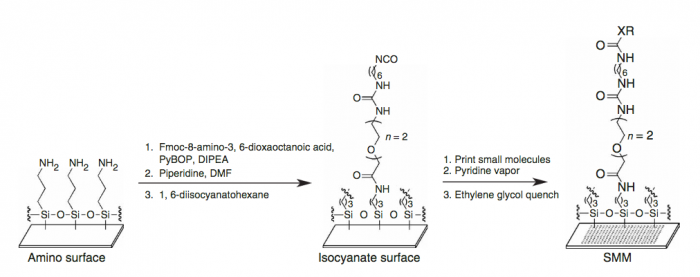
Our compound library is dissolved in DMSO and stored in 384-well plates. To dispense the compounds onto our functionalized glass slide, a robotic arm with a set of 48 metal pins is used to transfer the compounds to the glass slide. Each metal pin has a small slit in the end, and capillary action is used to precisely withdraw and dispense consistent volumes. When the pins touch the glass slide, the compound in DMSO is dispensed into a small circle of approximately 160 micron in diameter. Each pin prints one compound in two different locations on each slide, and then the pins are washed in water and DMSO. This process is repeated for each compound, resulting in our final microarray. The microarray is divided into 48 subarrays, and each subarray corresponds to one pin and contains 256 discrete spots. Within each subarray, we print a set of fluorescent compounds in the shape of an ‘X’ so that we can precisely determine where each spot is printed. After the compounds react, we quench the surface so that no electrophiles remain. This results in our final microarray; a collection of approximately 12,000 discrete spots displaying one compound each.
(Written by Rob Wilson. For more information read Bradner, J. E., McPherson, O. M., and Koehler, A. N. (2006) A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Prot. 1:2344-2352. PMID: 17406478.)
Protocols
Part 1: Complete Orientation quiz
Complete the orientation quiz with your partner. Though you are working with your partner, each student should record all answers independently. If you disagree with your partner on an answer, you should write what you think is the correct answer on your quiz.
Part 2: Review SMM experimental details
For timing reasons, you will not perform an SMM experiment this semester. Instead you will expand on the results gathered by researchers in the Koehler Laboratory to test putative small molecule binders in a secondary assay. To ensure that you are familiar with the steps used in an SMM screen, review the procedure below.
Perform SMM screen
To prepare for the SMM experiment, dilute the concentrated TDP43-RRM12 protein in TBS-T.
In your laboratory notebook, complete the following:
- Calculate the volume of TDP43-RRM12 and of TBS-T required to prepare the protein for the SMM experiment.
- Use 400 μg / mL for the concentration of concentrated TDP43-RRM12.
- Total volume should = 7 mL.
- Final concentration of TDP43-RRM12 should = 0.5 μg/mL.
To ensure the steps included below are clear, please watch the video tutorial linked here: [SMM Screen]. The steps are detailed below so you can follow along!
- Obtain two printed SMM slides from the front laboratory bench.
- Only hold them at the barcoded end as you would otherwise disrupt the ligands that are printed on the surface.
- Record the barcode numbers of your slides in your laboratory notebook.
- Carefully place each slide, barcode facing up, into a well of a 4-well chamber dish using the slide forceps.
- Again, only hold the slide at the barcoded end.
- Add 3 mL of the diluted protein solution to the chambers that contain your slides.
- Pipet the diluted protein solution onto the barcoded end of the slides.
- Avoid generating bubbles as you add your diluted protein solution.
- Gently rock the 4-well chamber dish back and forth until the slides are completely covered with the diluted protein solution.
- Cover the 4-well chamber dish with aluminum foil and incubate on the rocking table for 1 hr.
- During all incubations it is important to keep the slides covered as the fluorescein spots used for alignment during scanning are sensitive to light.
- Carefully pour the diluted protein solution from your 4-well chamber dish into a waste container by tipping the dish such that the liquid leaves the dish from the corner.
- The surface tension should keep the slides in the 4-well chamber dish; however, it is good to be careful that they do not fall into the waste container.
- Add ~3 mL of TBS-T to the chambers that contain your slides.
- Remember, pipet the liquid onto the barcoded end of the slides.
- Don't 'push' the liquid across the surface of the slide with the pipet. Instead, gently rock the 4-well chamber dish back and forth to completely cover your slides.
- Incubate the slides on the rocking table for 2 min.
- Don't forget to cover the 4-well chamber dish with aluminum foil.
- Pour the TBS-T from the 4-well chamber dish as above into a waste container.
- Complete Steps #8 - 10 a total of 3 times.
- Move your slides to a fresh 4-well chamber dish to eliminate carryover from the protein and/or buffer solutions.
- Add ~3 mL of TBS (not TBS-T!) to remove excess detergent and incubate your covered 4-well chamber dish on the rocking table for 2 min, then pour liquid into a waste container.
- Lastly, wash the slides with dH2O.
- Obtain 2 reservoirs and add fresh dH2O to each.
- Label the reservoirs '1' and '2'.
- Wash each slide by removing it from the 4-well chamber dish with the slide forceps and dunking it 8 times into reservoir 1, then dunking it 8 times into reservoir 2.
- Dry your slides by carefully insert your slides in the 'slide spinner' at the front laboratory bench.
- Be mindful not to scrape the face of your slides on the slide holder.
- Centrifuge your slides for 30 sec.
- Transfer your slides into fresh 50 mL conical tubes and wrap in aluminum foil.
- Store SMM slides at 4 °C until imaging.
Scan SMM slides
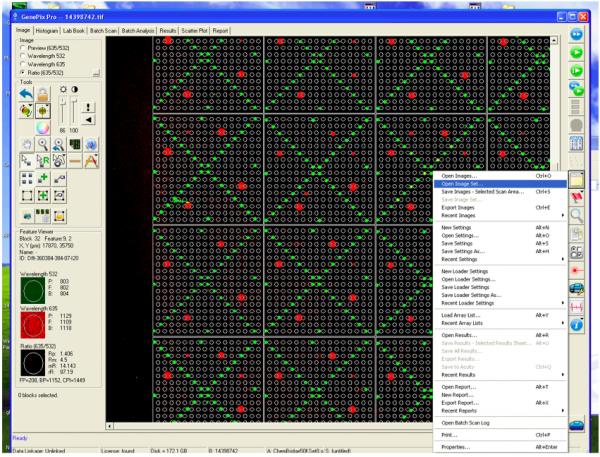
To check which of the small molecules, if any, may be able to bind TDP43-RRM12, the slides were imaged using a Genepix microarray scanner. The scanner measured the fluorescence signal emitted from the slide at two wavelengths: 532 nm and 635 nm. These images are the raw data that will be used to identify putative small molecule binders.
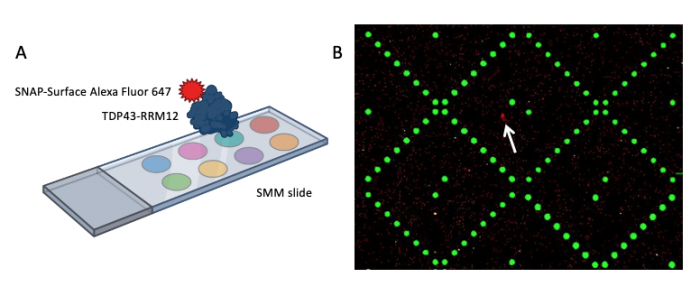
When the SMM slides are imaged, the scanner exposes each slide to excitation light specific to the fluorophores used in the experiment. As shown in the figure above, two fluorophores were used to evaluate small molecule binding to TDP43-RRM12. The green spots represent locations on the SMM slide where fluorescein was printed. Fluorescein is a fluorescent dye that emits at 532 nm and is used for alignment purposes. Correct alignment is critical to knowing which small molecules are in which spots on the slide. Red spots are indicative of small molecules that are bound by TDP43-RRM12. The signal is because the Alexa Fluor 647 emits at 635 nm.
During imaging, the scanner detects the intensity of the emitted fluorescence light to generate an image that can be analyzed. The intensity and position of the 'red' signal associated with putative small molecule binders is what will be assessed in the next laboratory session. This analysis will provide a list of 'hits' that can be used in future studies to identify drug candidates.
To ensure you are familiar with the steps involved in imaging the SMM experiment, please watch the video tutorial provided by a researcher from the Koehler Laboratory linked here: SMM Scanning. The steps are detailed below so you can follow along!
- Place the slide in the scanner barcode side down.
- Set the desired wavelengths that you want to scan.
- For fluorescein: 532 nm
- For Alexa Fluor 647: 635 nm
- Set the wavelength, filter, PMT, and power settings.
- Run a preview scan to optimize the PMT for the 635 nm (red) emission.
- Complete a full scan using the optimal PMT value.
- Following the scan, you will see an image of your slide as below.
In your laboratory notebook, complete the following:
- What color is emitted by fluorescein? What does this indicate on the scan (what is present in spots that emit this signal)?
- What color is emitted by Alexa Fluor 647? What does this indicate on the scan (what is present in spots that emit this signal)?
Reagents list
- small-molecule microarray slides (a gift from Koehler Laboratory)
- Tris-HCl buffered saline (TBS): 50 mM Tris-Hcl (pH = 7.5), 150 mM NaCl (from BioRad)
- TBS containing 0.1% Tween20 (TBS-T) (from BioRad)
- Genepix 4300 microarray scanner (from Molecular Devices)
- Genepix Pro software (from Molecular Devices)