Difference between revisions of "20.109(S21):M1D7"
(→Introduction:Analyze ligand titration curves) |
(→Protocols) |
||
| Line 35: | Line 35: | ||
'''Analyze scFv Flow Cytometry Titration Samples in FlowJo''' | '''Analyze scFv Flow Cytometry Titration Samples in FlowJo''' | ||
| − | #Access the flow cytometry data [https://www.dropbox.com/sh/1b7do6yu7zxs6cs/AABYY1nDhUd3Fr8hPfyCbL-6a?dl=0 here.] | + | #Access the flow cytometry data [https://www.dropbox.com/sh/1b7do6yu7zxs6cs/AABYY1nDhUd3Fr8hPfyCbL-6a?dl=0 here.] You and your team mate(s) will analyze the lysozyme titration data for the two clones you analyzed on "sequencing analysis day" or M1D5. You will also analyze the parental lysozyme titration data for comparison. |
#When you have activated your trial, a blank workspace should have opened up in FlowJo. If not, open the FlowJo_v10.7 program and create a new workspace by clicking the “new” button. | #When you have activated your trial, a blank workspace should have opened up in FlowJo. If not, open the FlowJo_v10.7 program and create a new workspace by clicking the “new” button. | ||
#Add '''all''' of the sample files to your workspace (FlowJo Tab --> Add samples) | #Add '''all''' of the sample files to your workspace (FlowJo Tab --> Add samples) | ||
| Line 43: | Line 43: | ||
#*The FL1-A channel and x-axis will measure the fluorescence intensity of the AF488 marker on our cells. Thus, dots farther to the right on the x axis represent cells that displayed more scFv on their surface. | #*The FL1-A channel and x-axis will measure the fluorescence intensity of the AF488 marker on our cells. Thus, dots farther to the right on the x axis represent cells that displayed more scFv on their surface. | ||
#*The FL4-A channel and y-axis will measure the fluorescence intensity of the AF647 marker on our cells. Thus, dots higher on the y axis represent cells with scFvs that bound lysozyme. | #*The FL4-A channel and y-axis will measure the fluorescence intensity of the AF647 marker on our cells. Thus, dots higher on the y axis represent cells with scFvs that bound lysozyme. | ||
| − | #We will now set two gates: yeast that displayed scFvs and yeast that did not display scFv. Despite the induction with galactose media not all yeast cells will display scFv on their surface (ex. Dead cells, budding cells). We want to analyze only the cells that displayed scFvs on their surface (right population) | + | #We will now set two gates: yeast that displayed scFvs and yeast that did not display scFv. Despite the induction with galactose media not all yeast cells will display scFv on their surface (ex. Dead cells, budding cells). We want to analyze only the cells that displayed scFvs on their surface (right population.) |
| − | #*Use the rectangle gating tool at the top to make a “displaying | + | #*Use the rectangle gating tool at the top to make a “displaying yeast” population. Your plot should look like Figure 2 below: <p>[[Image:Sp21 displaying yeast.jpeg|thumb|right|200px|Gating scFv Displaying Yeast]]</p> |
#We will now add statistics to our graph to obtain the '''Median Fluorescence Intensity''' of the y-axis AF647 binding signal. Under the “Edit” tab, choose “Add a Statistic”. A pop-up menu will appear. | #We will now add statistics to our graph to obtain the '''Median Fluorescence Intensity''' of the y-axis AF647 binding signal. Under the “Edit” tab, choose “Add a Statistic”. A pop-up menu will appear. | ||
#*Choose the “displaying yeast” as your population drop-down option. | #*Choose the “displaying yeast” as your population drop-down option. | ||
#*Choose “Median” in the statistic options menu on the left. | #*Choose “Median” in the statistic options menu on the left. | ||
#*Choose “FL4-A::FL4-A” as the statistic options menu on the left and select “Add” at the bottom. | #*Choose “FL4-A::FL4-A” as the statistic options menu on the left and select “Add” at the bottom. | ||
| − | |||
#Close the graph and return to the workspace screen. Your first sample should now have the following sub-populations listed below: <p>[[Image:Sp21 Median.jpeg|thumb|none|350px|Copy gates to other titration experiments]]</p> | #Close the graph and return to the workspace screen. Your first sample should now have the following sub-populations listed below: <p>[[Image:Sp21 Median.jpeg|thumb|none|350px|Copy gates to other titration experiments]]</p> | ||
#We now need to copy all of the '''sub-populations and statistics''' below the first sample to the other samples in our workspace. Select all of the subpopulations and statistics (select top, hold shift key, select bottom) and copy them (via ctrl+c, or copy in edit tab). Select the remaining samples and paste the subpopulations and statistics. All of the samples should now have the same five sub-populations and statistics. | #We now need to copy all of the '''sub-populations and statistics''' below the first sample to the other samples in our workspace. Select all of the subpopulations and statistics (select top, hold shift key, select bottom) and copy them (via ctrl+c, or copy in edit tab). Select the remaining samples and paste the subpopulations and statistics. All of the samples should now have the same five sub-populations and statistics. | ||
| − | #At this point, we need to check our control samples to ensure our gates are set correctly | + | #At this point, we need to check our control samples to ensure our gates are set correctly. We want to make sure that our "displaying yeast" gate excludes cells that do not display scFvs but may be slightly sticky to AF488. Make sure to check your controls. We want to set our gate to include minimal sticky cells. |
#Next, we will export the statistics in a table. Under the “FlowJo” tab, select “Table Editor” | #Next, we will export the statistics in a table. Under the “FlowJo” tab, select “Table Editor” | ||
#* Under the “Edit” tab in the Table Editor pop up window, select “Add Column”. | #* Under the “Edit” tab in the Table Editor pop up window, select “Add Column”. | ||
| − | #*Add the two statistics to your table that you just added to your samples in your workspace using the same populations (displaying | + | #*Add the two statistics to your table that you just added to your samples in your workspace using the same populations (displaying yeast), statistics (Median), and parameter (FL4-A::FL4-A). Your window should look like the following: <p>[[Image:Sp21 TableEditor.jpeg|thumb|none|550px|View of table editor]]</p> |
#*Return to the “Table Editor” tab and “Create Table” by pressing the gear symbol. | #*Return to the “Table Editor” tab and “Create Table” by pressing the gear symbol. | ||
| − | #*Save the table as an excel file for further analysis. | + | #*Save the table as an excel file for further analysis. These values are critical for next steps. |
<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| − | # | + | # Note the mutant number and annotate the mutations associated with each mutant as "Heavy Chain aaGly to Cys." |
| + | # Copy and paste the MFI values for the parental control, Mutant 1 and Mutant 2 your team analyzed in your notebook and on the discussion tab of this wiki page. | ||
| + | # What can you infer from the MFI values of your mutants vs. parental? Are the MFIs higher or lower for a given lysozyme concentration? | ||
| + | # What can you infer from the scatterplot titration? Do you think your mutants were good binders compared to the parental? | ||
=== Part 2: Plot titration binding curves via Matlab === | === Part 2: Plot titration binding curves via Matlab === | ||
| Line 69: | Line 71: | ||
# Open the Matlab code here: [[media: SolveKd_MultipleClones.m |Solve for Kd]] | # Open the Matlab code here: [[media: SolveKd_MultipleClones.m |Solve for Kd]] | ||
# | # | ||
| + | |||
<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
#Report the <math>K_d</math> of each of the clones from your graph. | #Report the <math>K_d</math> of each of the clones from your graph. | ||
| − | #*Which | + | #*Which mutant (or parental) has the highest <math>K_d</math>? |
| − | #*Which | + | #*Which mutant (or parental) has the lowest <math>K_d</math>? |
#*Rank the scFvs in order from weakest to strongest lysozyme binders. | #*Rank the scFvs in order from weakest to strongest lysozyme binders. | ||
| + | #Can you make any connections between the type of amino acid mutations and the strength of binding to lysozyme? | ||
==Reagents list== | ==Reagents list== | ||
Revision as of 02:52, 16 March 2021
Contents
Introduction:Analyze ligand titration curves
"Each protein in a cell collides with a tremendous variety of other molecules every second, but the great majority of these interactions are fleeting and have no functional consequences. However, in a small minority of these collisions, a more persistent interaction occurs between the protein and its ligand, docking the two into a precise relative orientation. This complex may stay assembled anywhere from seconds to days and perform a variety of functions: catalysis of a chemical reaction, construction of an intracellular structure or compartment, or transmission of signaling information from the environment or between intracellular compartments."
excerpt from Quantitative Fundamentals of Molecular and Cellular Bioengineering by K. Dane Wittrup, , Bruce Tidor, , Benjamin J. Hackel, , and Casim A. Sarkar. MIT Press, 2020.
How do we measure non-covalent binding between two molecules? The antigen, lysozyme (L), and antibody (Ab) form a complex (C), which can be written
$ L + Ab \rightleftharpoons\ C $
At equilibrium, the rates of the forward reaction (rate constant = $ k_f $) and reverse reaction (rate constant = $ k_r $) must be equivalent. Solving this equivalence yields an equilibrium dissociation constant $ K_d $, which may be defined either as $ k_r/k_f $, or as $ [Ab][L]/[C] $, where brackets indicate the molar concentration of a species.
Meanwhile, the fraction of antibody that are bound to antigen at equilibrium, often called y or θ, is $ C/Ab_{TOT} $, where $ Ab_{TOT} $ indicates total (both bound and unbound) receptors. Note that the position of the equilibrium (i.e., y) depends on the starting concentrations of the reactants; however, $ K_d $ is always the same value. The total number of antibody $ Ab_{TOT} $= [C] (L-bound Ab) + [Ab] (unbound Ab).
$ \qquad y = {[C] \over Ab_{TOT}} \qquad = \qquad {[C] \over [C] + [Ab]} \qquad = \qquad {[L] \over [L] + [K_d]} \qquad $
where the right-hand equation was derived by algebraic substitution. If the antigen concentration is in excess of the concentration of the antibody, [L] may be approximated as a constant, L, for any given equilibrium.
Protocols
Motivation: To calculate the apparent $ K_d $ from our lysozyme titration experiments we will analyze the flow cytometry scatterplot data collected from the Accuri C6.
Part 1: Analyze flow cytometry scatter plots using FlowJo and generate Median Fluorescent Intensity (MFI) table
Today, we will be analyzing the flow cytometry data that we collected last lab class in order to assess the binding of your mutant scFv clones to lysozyme as compared to the parental scFv Lyso_scFv_6. On M1D5, you compared the sequences of two different mutant clones to the parental scFv DNA. This parental clone is reported to have a $ K_d $ of of 6 nM for binding to lysozyme. To assess differences in binding affinity between our mutants and the parental clone we will calculate and compare the apparent $ K_d $ of two mutants and the parental clone. Download FlowJo on to your computer and activate free 30-day trial
- Go to https://www.flowjo.com/solutions/flowjo/free-trial and click “Download”. Complete the software download instructions to install FlowJo v10.7 for your system (Mac or Windows).
- Once the software installation has finished, open the program and locate your computer’s hardware ID number in the license pop-up window. Complete the form using your email and ID number at the above link. If the license pop-up window does not automatically appear with your ID number, instructions to find the number through FlowJo are listed here: https://docs.flowjo.com/flowjo/faq/general-faq/locating-hwa/.
- Use the serial number sent to your email to activate your 30-day free trial.
Analyze scFv Flow Cytometry Titration Samples in FlowJo
- Access the flow cytometry data here. You and your team mate(s) will analyze the lysozyme titration data for the two clones you analyzed on "sequencing analysis day" or M1D5. You will also analyze the parental lysozyme titration data for comparison.
- When you have activated your trial, a blank workspace should have opened up in FlowJo. If not, open the FlowJo_v10.7 program and create a new workspace by clicking the “new” button.
- Add all of the sample files to your workspace (FlowJo Tab --> Add samples)
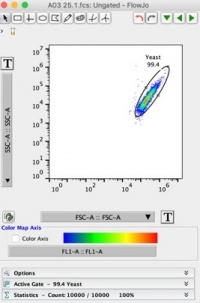
- Click on the first sample to open FSC-A vs. SSC-A plot. Change the axis to log scale by pressing the large [T] buttons by the axis labels. Use the oval gating tool at the top to select yeast cells (and exclude extraneous particles). Label this population as “yeast”. Your plot should look like Figure 1 below:
- Close the graph window and go back to the main workspace page and click on now gated “yeast” population below the first sample. This will open up a new graph with only the gated cells.
- We will now change our axis on the yeast scatterplot to analyze the data based on the fluorescence markers used in the secondary staining. Change the x-axis of the plot from “FSC-A::FSC-A” to “FL1-A::FL1-A” and the y-axis of the plot from “SSC-A::SSC-A” to “FL4-A::FL4-A”.
- The FL1-A channel and x-axis will measure the fluorescence intensity of the AF488 marker on our cells. Thus, dots farther to the right on the x axis represent cells that displayed more scFv on their surface.
- The FL4-A channel and y-axis will measure the fluorescence intensity of the AF647 marker on our cells. Thus, dots higher on the y axis represent cells with scFvs that bound lysozyme.
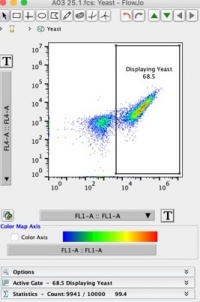
- We will now set two gates: yeast that displayed scFvs and yeast that did not display scFv. Despite the induction with galactose media not all yeast cells will display scFv on their surface (ex. Dead cells, budding cells). We want to analyze only the cells that displayed scFvs on their surface (right population.)
- Use the rectangle gating tool at the top to make a “displaying yeast” population. Your plot should look like Figure 2 below:
- We will now add statistics to our graph to obtain the Median Fluorescence Intensity of the y-axis AF647 binding signal. Under the “Edit” tab, choose “Add a Statistic”. A pop-up menu will appear.
- Choose the “displaying yeast” as your population drop-down option.
- Choose “Median” in the statistic options menu on the left.
- Choose “FL4-A::FL4-A” as the statistic options menu on the left and select “Add” at the bottom.
- Close the graph and return to the workspace screen. Your first sample should now have the following sub-populations listed below:
- We now need to copy all of the sub-populations and statistics below the first sample to the other samples in our workspace. Select all of the subpopulations and statistics (select top, hold shift key, select bottom) and copy them (via ctrl+c, or copy in edit tab). Select the remaining samples and paste the subpopulations and statistics. All of the samples should now have the same five sub-populations and statistics.
- At this point, we need to check our control samples to ensure our gates are set correctly. We want to make sure that our "displaying yeast" gate excludes cells that do not display scFvs but may be slightly sticky to AF488. Make sure to check your controls. We want to set our gate to include minimal sticky cells.
- Next, we will export the statistics in a table. Under the “FlowJo” tab, select “Table Editor”
- Under the “Edit” tab in the Table Editor pop up window, select “Add Column”.
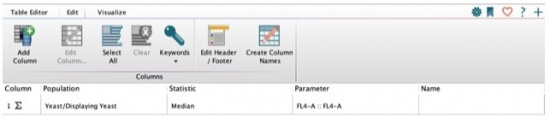
- Add the two statistics to your table that you just added to your samples in your workspace using the same populations (displaying yeast), statistics (Median), and parameter (FL4-A::FL4-A). Your window should look like the following:
- Return to the “Table Editor” tab and “Create Table” by pressing the gear symbol.
- Save the table as an excel file for further analysis. These values are critical for next steps.
In your laboratory notebook, complete the following:
- Note the mutant number and annotate the mutations associated with each mutant as "Heavy Chain aaGly to Cys."
- Copy and paste the MFI values for the parental control, Mutant 1 and Mutant 2 your team analyzed in your notebook and on the discussion tab of this wiki page.
- What can you infer from the MFI values of your mutants vs. parental? Are the MFIs higher or lower for a given lysozyme concentration?
- What can you infer from the scatterplot titration? Do you think your mutants were good binders compared to the parental?
Part 2: Plot titration binding curves via Matlab
Motivation: To calculate the apparent $ K_d $ from the median fluorescent intensity of our flow cytometry titration we will use Matlab to carry out the statistical analysis and fit a curve to the data.
You will need to either run Matlab software on your computer, or by using the online Matlab server found here: Matlab Online. Please note that you will need to make an account with Matlab using your MIT email address and use the MIT software license for free access. Instructions below are for Matlab online.
- Open the Matlab code here: Solve for Kd
In your laboratory notebook, complete the following:
- Report the $ K_d $ of each of the clones from your graph.
- Which mutant (or parental) has the highest $ K_d $?
- Which mutant (or parental) has the lowest $ K_d $?
- Rank the scFvs in order from weakest to strongest lysozyme binders.
- Can you make any connections between the type of amino acid mutations and the strength of binding to lysozyme?
Reagents list
Next day: Perform protein purification protocol