Difference between revisions of "20.109(S16):Purify protein (Day6)"
Noreen Lyell (Talk | contribs) (→Part 3A: Nickel-agarose purification) |
Noreen Lyell (Talk | contribs) (→Introduction) |
||
| Line 13: | Line 13: | ||
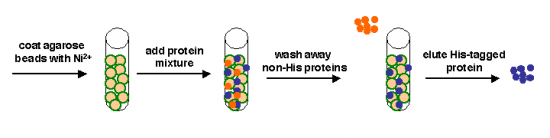
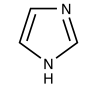
Today we will use a nickel-agarose resin to separate our protein of interest (IPC) from the other proteins present in the bacteria. The His-tagged protein will preferentially bind to the nickel-coated beads, while proteins irrelevant to our purposes in Module 1 can be washed away. Remember, the BL21(DE3)pLysS cells are not only producing our protein, but also the proteins needed for cellular function and survival. Finally, a high concentration of imidazole (which is the side chain of histidine) can be used to elute the His-tagged inverse pericam by competition. Due to the inherent fragility of IPC, we will add several components to our protein extraction and purification reagents: bovine serum albumin (BSA), which is a protein stabilizer, and a cocktail of protease inhibitors. | Today we will use a nickel-agarose resin to separate our protein of interest (IPC) from the other proteins present in the bacteria. The His-tagged protein will preferentially bind to the nickel-coated beads, while proteins irrelevant to our purposes in Module 1 can be washed away. Remember, the BL21(DE3)pLysS cells are not only producing our protein, but also the proteins needed for cellular function and survival. Finally, a high concentration of imidazole (which is the side chain of histidine) can be used to elute the His-tagged inverse pericam by competition. Due to the inherent fragility of IPC, we will add several components to our protein extraction and purification reagents: bovine serum albumin (BSA), which is a protein stabilizer, and a cocktail of protease inhibitors. | ||
| − | [[Image:20.109_Histidine.png|thumb| | + | [[Image:20.109_Histidine.png|thumb|200px|right|'''Histidine''']] |
| − | [[Image:20.109_Imidazole.png|thumb| | + | [[Image:20.109_Imidazole.png|thumb|100px|right|'''Imidazole''']] |
Prior to purifying our protein, we will lyse the bacteria and save the whole cell extracts to later run on a protein gel. This procedure is called SDS-PAGE, for sodium dodecyl sulfate-polyacrylamide gel electrophoresis. SDS is an ionic surfactant (or detergent), which denatures the proteins and coats them with a negative charge. Since denatured proteins are linear, they will move through the gel at a speed inversely proportional to their molecular weight, just like DNA on agarose gels. (Non-denatured proteins run according to their molecular weight, shape, and charge.) As we did with DNA gels, we will run a reference ladder containing proteins of known molecular weight and amount. When running –IPTG and +IPTG samples side-by-side, you should see a protein band at the expected molecular weight for inverse pericam (47 kDa + 3 kDa from the His-tag = 50 kDa), which may be very faint or non-existent in the -IPTG control sample, but bright and thick in the +IPTG induced sample. To visualize all the proteins released by the bacteria, you will stain the gels with Coomassie Brilliant Blue (actually, a variant called BioSafe Coomassie). This is a non-specific stain for all proteins. | Prior to purifying our protein, we will lyse the bacteria and save the whole cell extracts to later run on a protein gel. This procedure is called SDS-PAGE, for sodium dodecyl sulfate-polyacrylamide gel electrophoresis. SDS is an ionic surfactant (or detergent), which denatures the proteins and coats them with a negative charge. Since denatured proteins are linear, they will move through the gel at a speed inversely proportional to their molecular weight, just like DNA on agarose gels. (Non-denatured proteins run according to their molecular weight, shape, and charge.) As we did with DNA gels, we will run a reference ladder containing proteins of known molecular weight and amount. When running –IPTG and +IPTG samples side-by-side, you should see a protein band at the expected molecular weight for inverse pericam (47 kDa + 3 kDa from the His-tag = 50 kDa), which may be very faint or non-existent in the -IPTG control sample, but bright and thick in the +IPTG induced sample. To visualize all the proteins released by the bacteria, you will stain the gels with Coomassie Brilliant Blue (actually, a variant called BioSafe Coomassie). This is a non-specific stain for all proteins. | ||
Revision as of 14:46, 1 February 2016
Introduction
Last time you used the lactose-analogue IPTG to induce expression of inverse pericam in BL21(DE3)pLysS bacteria. Today you will isolate your mutated IPC and the wild-type IPC from the bacterial cells. You will also begin characterizing your wild-type and mutant proteins.
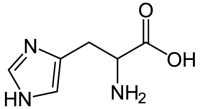
The bacterial expression vector we are using (pRSET) contains six histidine codons downstream of a bacterial promoter and in-frame with a start codon. Our resultant protein is therefore marked by the presence of these additional encoded residues, or His-tagged. Histidine has several interesting properties, notably its near-neutral pKa, and His-rich peptides are promiscuous binders, particularly to metals. (For example, histidine side chains help coordinate iron molecules in hemoglobin.)
Today we will use a nickel-agarose resin to separate our protein of interest (IPC) from the other proteins present in the bacteria. The His-tagged protein will preferentially bind to the nickel-coated beads, while proteins irrelevant to our purposes in Module 1 can be washed away. Remember, the BL21(DE3)pLysS cells are not only producing our protein, but also the proteins needed for cellular function and survival. Finally, a high concentration of imidazole (which is the side chain of histidine) can be used to elute the His-tagged inverse pericam by competition. Due to the inherent fragility of IPC, we will add several components to our protein extraction and purification reagents: bovine serum albumin (BSA), which is a protein stabilizer, and a cocktail of protease inhibitors.
Prior to purifying our protein, we will lyse the bacteria and save the whole cell extracts to later run on a protein gel. This procedure is called SDS-PAGE, for sodium dodecyl sulfate-polyacrylamide gel electrophoresis. SDS is an ionic surfactant (or detergent), which denatures the proteins and coats them with a negative charge. Since denatured proteins are linear, they will move through the gel at a speed inversely proportional to their molecular weight, just like DNA on agarose gels. (Non-denatured proteins run according to their molecular weight, shape, and charge.) As we did with DNA gels, we will run a reference ladder containing proteins of known molecular weight and amount. When running –IPTG and +IPTG samples side-by-side, you should see a protein band at the expected molecular weight for inverse pericam (47 kDa + 3 kDa from the His-tag = 50 kDa), which may be very faint or non-existent in the -IPTG control sample, but bright and thick in the +IPTG induced sample. To visualize all the proteins released by the bacteria, you will stain the gels with Coomassie Brilliant Blue (actually, a variant called BioSafe Coomassie). This is a non-specific stain for all proteins.
After purifying inverse pericam from your bacterial lysates, you will measure the protein concentration by the Bradford colorimetric assay, named after the scientist who first published it. The dye used in this assay is the same one you will use to stain your protein gels – Coomassie. In acidic solution, Coomassie normally has an absorbance peak at ~465 nm (blue light, so solution appears dark red), but this peak is shifted to 595 nm (orange light, so solution turns to blue) upon binding to protein. Protein binding occurs primarily via arginine, as well as other basic and aromatic residues, as described here. The concentration of protein present in a sample is thus proportional to the 595 nm absorbance peak, and its absolute value can be determined using a standard curve of reference protein. We do not have a sample of inverse pericam with a known quantity of protein, so today we will use BSA as a reference protein. Because the compositions of IPC and BSA with respect to arginine may vary, this assay will really only give the relative concentrations of your protein samples, and the absolute concentrations will have an associated error.
Protocols
Part 1: Lysis of cells producing wild-type and mutant IPC
- You will be given a 3 mL aliquot of room temperature BugBuster buffer (bacterial lysis and protein extraction solution), which contains 0.1% bovine serum albumin (BSA, a stabilizer), a nuclease to degrade nucleic acids, and a protease inhibitor cocktail to guard against protein degradation.
- When you are ready to begin, add 1:1000 of cold lysing enzyme mixture (obtained from teaching staff) to the lysis solution.
- Per cell pellet (4 total), add the appropriate volume of enzyme-containing BugBuster and resuspend by pipetting until the solution is relatively homogeneous.
- Resuspend -IPTG samples in 300 μL, and +IPTG samples in 600 μL - do you remember why?
- Pipet up and down to mix.
- Incubate the solutions (at room temperature) for 10 minutes on the nutator.
- During this incubation, you may begin the resin preparation described in Part 3.
- Finally, centrifuge for 10 minutes at maximum speed and transfer supernatants to fresh tubes.
- While one partner completes Part 2, the other partner can begin/continue with the resin preparation in Part 3.
Part 2: Advance preparation for SDS-PAGE of protein extracts
- Last time you measured the amount of cells in each of your samples (-IPTG and +IPTG of the wild-type IPC and one correct mutant). (If you ran cultures overnight, the teaching faculty measured the +IPTG samples for you and posted the results.) Look back at your measurements, and find the sample with the lowest cell concentration. Set aside 15 μL of this sample for PAGE analysis in an eppendorf.
- For your other three samples, you should take the amount of bacterial lysate corresponding to the same number of cells as the lowest concentration sample. For example, if the OD600 of your WT -IPTG sample was 0.05, and the OD600 of your WT +IPTG sample was 0.30, you would take 15 μL of the -IPTG, but only 2.5 μL of the +IPTG sample.
- Next, add enough water so the each sample has 15 μL of liquid in it. You might use the table below to guide your work.
- Finally, add 3 μL of 6X sample buffer to 15 μL of each of your diluted lysates. These will be stored in the freezer until next time.
| Sample Name | OD600 | Sample Volume (μL) | Water Volume (μL) | Total Volume (μL) |
|---|---|---|---|---|
| -IPTG WT | 15 | |||
| +IPTG WT | 15 | |||
| -IPTG mutant | 15 | |||
| +IPTG mutant | 15 |
Part 3: Protein purification
Part 3A: Nickel-agarose purification
You will process two samples (+IPTG wild-type and +IPTG mutant IPC) according to the following procedure. Keep all buffers on ice when not in use. All spins should be performed at 1000 rcf (3300 rpm) for 1 minute.
- The following buffers are aliquoted and located at the front bench:
- 1X Ni-NTA Bind Buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 10 mM imidazole)
- 1X Ni-NTA Wash Buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 20 mM imidazole)
- 1X Ni-NTA Elute Buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 250 mM imidazole)
- Note: Two special waste streams should be created for this affinity purification procedure, (1) nickel waste for the 50% slurry, and (2) imidazole waste for the Bind, Wash, and Elute Buffers.
- Gently mix the Ni-charged agarose resin to fully resuspend it, then distribute 400 μL of the 50% slurry to each of two 2 mL centrifuge tubes.
- Label one tube as wild type and the other as mutant.
- Add 1.6 mL (2 x 800 μL) of 1X Ni-NTA Bind Buffer to the resin.
- Resuspend the slurry by pippeting the solution up and down several times (10-15), then centrifuge at 3000 g for 1 minute.
- Carefully remove the supernatent and discard it in the appropriate waste stream.
- Add your cleared cell lysate from Part 1 to the resin, then put your tubes on the nutator at 4°C for 30 minutes.
- Be sure to add the wild type and mutant lysates to the correct tubes!
- Centrifuge at 3000 g for 1 minute.
- Remove the supernatent and discard it in the appropriate waste stream.
- Add 1 mL of 1X Ni-NTA Wash Buffer to the resin.
- Centrifuge at 3000 g for 1 minute.
- Remove the supernatent and discard it in the appropriate waste stream.
- Repeat Steps #8-10.
- Finally, you will collect your protein. Add 75 μL of Elute Buffer, resuspend, and spin as usual. Do not throw away the supernatant! Instead, transfer it to a fresh eppendorf tube, labeled “pure IPC X#Z" or “pure IPC WT.”
- Do not throw away the resin yet either! Instead, repeat Step #12 two more times and add the supernatent to the sample collected in Step #12.
Part 3B: Desalting
We found from pilot studies that imidazole affects the binding curves of inverse pericams. Thus, you will continue purifying your proteins by removing any low molecular weight compounds.
- For each of your two samples, snap off the bottom of a Zeba column, place in a 15 mL conical tube, and loosen the column's cap.
- In the large centrifuge across from the freezer, spin your columns at 1000 rcf (which is 2100 rpm for the rotor inside this centrifuge) for 2 minutes.
- The timing function on this centrifuge does not work! Bring your timer and manually turn the centrifuge off after 2 minutes. Start the timer once the rpm value approaches 2000.
- Because we all have to share one centrifuge, ideally spin with at least 2 other groups.
- When you remove your columns, the resin inside should be slanted. Make a mark right on the column where the highest point of the resin occurs, and orient that mark facing outward in the next step.
- Transfer the column to a fresh 15 mL conical tube, and then gently apply your ~1 mL of protein to the center of the compacted resin.
- Repeat the 2-minute spin step just as before.
- Immediately after eluting your protein, transfer 10 μL of purified protein to a clean eppendorf tube for assaying protein concentrations (Part 4) and 15 μL to a separate eppendorf tube for SDS-PAGE analysis (give this tube to your instructor).
- Then add a 1:100 dilution of 10% BSA to the remaining protein (10 μL of BSA for ~1 mL of protein).
Part 4: Protein concentration
- Prepare 12 mL Bradford reagent from the 5x concentrated stock by adding to a 9.6 mL water aliquot.
- Obtain BSA standards from the teaching faculty.
- Each tube already contains exactly 10 μL of standard (or plain water, for your blank solution).
- Add 1 mL of Bradford reagent to each standard, as well as to your two unknown protein samples (10 μL set aside in Part 3B). Incubate 10-20 minutes at room temperature.
- Measure the absorbance of each sample at 595 nm. Work as quickly as you can, because the absorbance will continue to slowly change over time. To get a sense of the error incurred due to the ongoing reaction, measure your BSA 0.1 mg/mL sample both at the beginning and at the end of your run.
| Sample |
A595 |
|---|---|
| Blank (at start) | |
| BSA 0.1 mg/mL (start) | |
| BSA 0.2 mg/mL | |
| BSA 0.4 mg/mL | |
| BSA 0.6 mg/mL | |
| BSA 0.8 mg/mL | |
| BSA 1.0 mg/mL | |
| WT IPC | |
| Mutant X#Y | |
| BSA 0.1 mg/mL (end) |
Reagent list
- Cell lysis
- EasyLyse from Epicentre Biotechnologies
- Bovine Serum Albumin
- Protease Inibitor Set, EDTA-Free from Calbiochem
- Lysis Enzyme from Epicentre Biotechnologies
- 6X Laemmli sample buffer from Boston BioProducts
- 2% SDS, 6% glycerol, 0.03% Bromophenol Blue in 375 mM Tris-HCl pH 6.8, + 9% β-mercaptoethanol
- Protein purification from Novagen/Calbiochem
- His-Bind Purification Kit buffers
- His-Bind Resin, Ni-Charged
- Zeba Desalt Spin Columns from Thermo Scientific
- 7000 Da MW cut-off
- Protein concentration
- Bio-Rad Protein Assay (Bradford Reagent)
Next day: Characterize protein expression