20.109(S16):In situ cloning (Day1)
Contents
Introduction
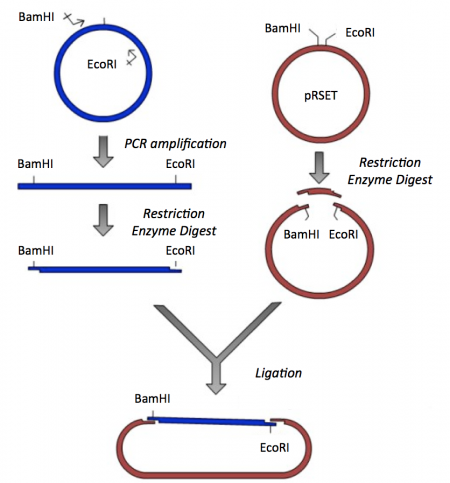
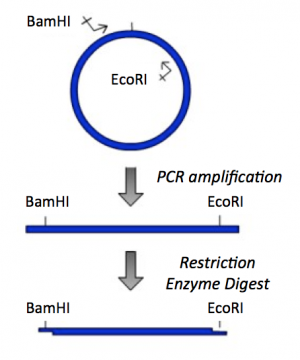
Though the theme of Module 1 is protein engineering, today will focus on a few key techniques used in DNA engineering. Because the sequence of proteins is determined by the sequence of the genes that encode them, learning how to manipulate DNA is an important first step. Today you will complete a cloning reaction to generate a protein expression vector that contains a gene that encodes a calcium-sensing protein. This process is illustrated in the schematic below. Later you will use this construct to engineer a new calcium-sensing protein.
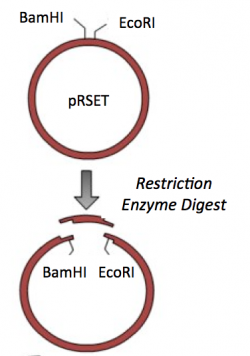
The cloning vector we will use is pRSET. This vector has several features that make it ideal for cloning and protein expression -- both of which are important for this module. The calcium-sensing protein we will study this module is inverse pericam (IPC). We will discuss this protein in much more detail later, for now it is sufficient to know that is used to measure calcium concentrations. To generate your final product you will use three common DNA engineering techniques: PCR amplification, restriction enzyme digestion, and ligation.
PCR amplification
The applications of PCR are widespread, from forensics to molecular biology to evolution, but the goal of any PCR is the same: to generate many copies of DNA from a single or a few specific sequence(s) (called the “target” or “template”).
In addition to the target, PCR requires only three components: primers to bind sequence flanking the target, dNTPs to polymerize, and a heat-stable polymerase to carry out the synthesis reaction over and over and over. PCR is a three-step process (denature, anneal, extend) and these steps are repeated 20 or more times. After 30 cycles of PCR, there could be as many as a billion copies of the original target sequence.
Based on the numerous applications of PCR, it may seem that the technique has been around forever. In fact it is just over 30 years old. In 1984, Kary Mullis described this technique for amplifying DNA of known or unknown sequence, realizing immediately the significance of his insight.
"Dear Thor!," I exclaimed. I had solved the most annoying problems in DNA chemistry in a single lightening bolt. Abundance and distinction. With two oligonucleotides, DNA polymerase, and the four nucleosidetriphosphates I could make as much of a DNA sequence as I wanted and I could make it on a fragment of a specific size that I could distinguish easily. Somehow, I thought, it had to be an illusion. Otherwise it would change DNA chemistry forever. Otherwise it would make me famous. It was too easy. Someone else would have done it and I would surely have heard of it. We would be doing it all the time. What was I failing to see? "Jennifer, wake up. I've thought of something incredible." --Kary Mullis from his Nobel lecture; December 8, 1983
Restriction enzyme digest
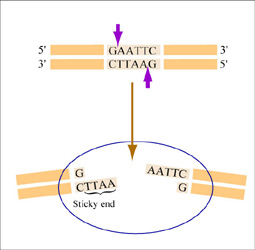
Restriction endonucleases, also called restriction enzymes, cut (“digest”) DNA at specific sequences of bases. The restriction enzymes are named according to the prokaryotic organism from which they were isolated. For example, the restriction endonuclease EcoRI (pronounced “echo-are-one”) was originally isolated from E. coli giving it the “Eco” part of the name. “RI” indicates the particular version on the E. coli strain (RY13) and the fact that it was the first restriction enzyme isolated from this strain.
The sequence of DNA that is bound and cleaved by an endonuclease is called the recognition sequence or restriction site. These sequences are usually four or six base pairs long and palindromic, that is, they read the same 5’ to 3’ on the top and bottom strand of DNA. For example, the recognition sequence for EcoRI (see also figure at right) is
5’ GAATTC 3’
3’ CTTAAG 5’
Unlike EcoRI, some other restriction enzymes cut precisely in the middle of the palindromic DNA sequence, thus leaving no overhangs after digestion. The single-stranded overhangs resulting from DNA digestion by enzymes such as EcoRI are called sticky ends, while double-stranded ends resulting from digestion by enzymes such as HaeIII are called blunt ends. HaeIII recognizes
5’ GGCC 3’
3’ CCGG 5’
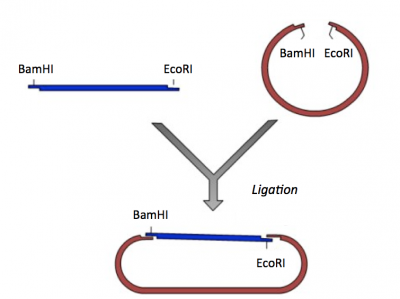
Ligation
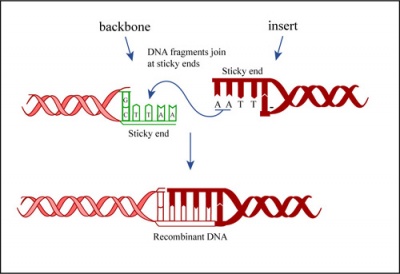
During the ligation reactions, hydrogen bonds will form between the overhangs on the fragments, and then the ligase will repair the phosphate backbone, creating a stable circular plasmid (as shown in the figure below).
Protocols
Because DNA engineering at the benchtop can take days, if not weeks, you will generate your clone in silico today. You can use any DNA manipulation software you choose to complete the protocols, but the instructions provided are for APE (A Plasmid Editor, created by M. Wayne Davis at the University of Utah). The software can be downloaded free-of-charge (add site) onto your personal computer or you can use the teaching laboratory computers. Please note that if you use a different program the teaching faculty may not be able to assist you.
Be sure to document your work and answer all questions in your notebook as you progress through the exercises below.
Part 1: PCR amplification and restriction enzyme digest of IPC insert
To amplify a specific sequence of DNA, you first need to design primers -- one primer that anneals to the start of the sequence and a second primer that anneals to the end of the sequence. Today you will design a 'forward' primer that reads toward the IPC gene and a 'reverse' primer that anneals to the opposite DNA strand at the end of the IPC gene and reads back into it. Each primer will consist of two parts. The 'landing sequence' will anneal to the gene and the 'flap sequence' will be used to add restriction enzyme recognition sequences to your IPC insert.
- Find the IPC sequence attached here.
- Open APE then copy and paste the sequence into a new window.
- Because we want to amplify the entire gene, the forward primer will begin with the first basepair of the sequence.
- Record the first 20 basepairs of the IPC gene sequence in your notebook.
- Visually inspect this sequence and make notes based on the following guidelines for primer design:
- Length: 17-28 basepairs
- GC Content: 50-60%
- Tm: 60-65°C
- Avoid hairpins, complementation between primers, repetitive sequences
- Several websites are available to help you evaluate your primers more accurately.
- Copy and paste the 20 basepair sequence into Sequence box at the IDT website.
- Leave the defaults for stems and loops as they are and then click Analyze.
- If you primer does not fit the guidelines provided above, try altering the length. Remember that the 5’ end of the landing sequence must not change or you will delete basepairs from your gene.
- When you are happy with the landing sequence, use the Features tool to mark the sequence on your APE file (Features → New Feature).
- Now that you have your landing sequence you will add a flap sequence that introduces a restriction enzyme recognition sequence.
- We will use EcoRI in our cloning strategy (recognition sequence = GAATTC).
- Add the recognition sequence for the EcoRI restriction enzyme to the landing sequence. Consider how PCR amplification occurs to determine which end of your primer should carry the flap sequence.
- In addition to the recognition sequence, it is important to include a 6 basepair 'tail' or 'junk' sequence to ensure the restriction enzyme is able to bind and cut the DNA. Add CATTAG as the junk sequence. Carefully consider where this sequence should appear in your primer.
- Record the sequence (5' - 3') of your forward primer in your notebook.
- Use steps 2-5 to design your reverse primer.
- Because you want to amplify the entire gene you should start with the last basepair of the sequence.
- Remember that the reverse primer anneals to the opposite DNA strand at the end of the IPC gene and reads back into it. Keep this in mind when you add the flap sequence and when you record the sequence of your primer in your notebook.
- Create a new APE file that depicts the IPC product you would expect if you used your primers in a PCR amplification reaction.
- What has been added to the sequence?
- Add the sequence information to your notebook (it may be easiest to screen capture your work station in APE and embed the image in your notebook).
- Now that you have your amplified IPC insert, you need to digest with EcoRI to generate 'sticky ends' to prepare your gene for the ligation step.
- Search the NEB catalog to where EcoRI cuts within its recognition sequence.
- Create another new APE file that depicts your amplified IPC product following an EcoRI digest.
- What had been removed from the sequence?
Part 2: Restriction enzyme digest of pRSET vector
Part 3: Ligation of IPC insert and pRSET vector
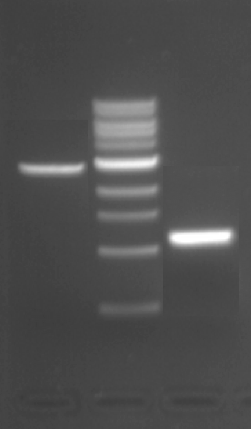
When completed a ligation at the bench, one important step is to calculate the amounts of DNA you will use in the reaction. Use the steps below to complete calculate the amount of IPC insert and pRSET vector you would use to complete this ligation in the laboratory.
- Separately calculate the concentration of backbone and of insert based on the recovery gel posted below. Refer to the NEB marker definitions to estimate the ng of DNA in each lane, and refer to your notebook/protocol for the relevant volume basis. Note that the ng listed are for 10 μL of ladder (and you loaded 20 μL of ladder).
- You may convert the mass concentration to a molar concentration, using the fact that a typical DNA base is 500 g/mol. This conversion will mostly cancel out between the insert and the backbone, except for the difference in number of bases. Feel free to either omit steps that will cancel if you are comfortable doing so, or to keep them if you follow the lab math better that way.
- Ideally, you will use 50-100 ng of backbone in the upcoming ligation. Referring to the mass concentration, what volume of DNA will this amount require?
- Ideally, you will use a 4:1 molar ratio of insert to backbone. Referring to the molar concentrations, how much insert do you need per μL of backbone?
- A 15 μL scale ligation should not include more than 13.5 μL of DNA. If your backbone and insert volumes total to greater than this amount, you must (1) scale down both DNA amounts, using less than 50 ng backbone and/or (2) stray from the ideal 4:1 molar ratio. You may ask the teaching faculty for advice during class if you are unsure what choice is best, but make and submit an initial ligation plan for now.
Part 4: Confirmation digest
Next day: Design mutation primers
Previous day: Orientation