20.109(S16):In situ cloning (Day1)
Contents
Introduction
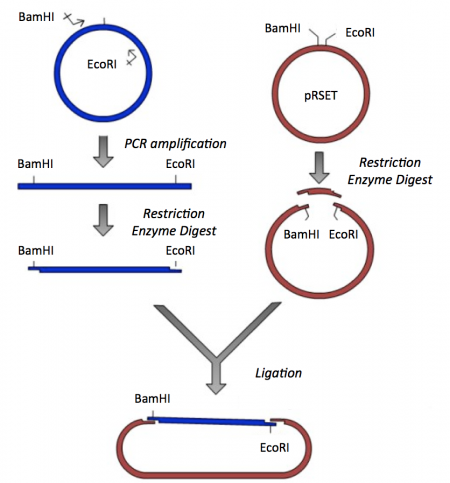
Though the theme of Module 1 is protein engineering, today will focus on a few key techniques used in DNA engineering. Because the sequence of proteins is determined by the sequence of the genes that encode them, learning how to manipulate DNA is an important first step. Today you will complete a cloning reaction to generate a protein expression vector that contains a gene that encodes a calcium-sensing protein. This process is illustrated in the schematic below. Later you will use this construct to engineer a new calcium-sensing protein.
The cloning vector we will use is pRSET. This vector has several features that make it ideal for cloning and protein expression -- both of which are important for this module. The calcium-sensing protein we will study this module is inverse pericam (IPC). We will discuss this protein in much more detail later, for now it is sufficient to know that is used to measure calcium concentrations. To generate your final product you will use three common DNA engineering techniques: PCR amplification, restriction enzyme digestion, and ligation.
PCR amplification
To begin your recombination study you will be using a method called the Polymerase Chain Reaction (PCR). The applications of PCR are widespread, from forensics to molecular biology to evolution, but the goal of any PCR is the same: to generate many copies of DNA from a single or a few specific sequence(s) (called the “target” or “template”).
In addition to the target, PCR requires only three components: primers to bind sequences flanking the target, dNTPs to polymerize, and a heat-stable polymerase to carry out the synthesis reaction over and over and over. PCR is a three-step process (denature, anneal, extend) and these steps are repeated 20 or more times. After 30 cycles of PCR, there could be as many as a billion copies of the original target sequence.
Based on the numerous applications of PCR, it may seem that the technique has been around forever. In fact it is just over 30 years old. In 1984, Kary Mullis described this technique for amplifying DNA of known or unknown sequence, realizing immediately the significance of his insight.
"Dear Thor!," I exclaimed. I had solved the most annoying problems in DNA chemistry in a single lightening bolt. Abundance and distinction. With two oligonucleotides, DNA polymerase, and the four nucleosidetriphosphates I could make as much of a DNA sequence as I wanted and I could make it on a fragment of a specific size that I could distinguish easily. Somehow, I thought, it had to be an illusion. Otherwise it would change DNA chemistry forever. Otherwise it would make me famous. It was too easy. Someone else would have done it and I would surely have heard of it. We would be doing it all the time. What was I failing to see? "Jennifer, wake up. I've thought of something incredible." --Kary Mullis from his Nobel lecture; December 8, 1983
Restriction enzyme digest
Ligation
Next day: Design mutation primers
Previous day: Orientation