20.109(S16):In situ cloning (Day1)
Contents
Introduction
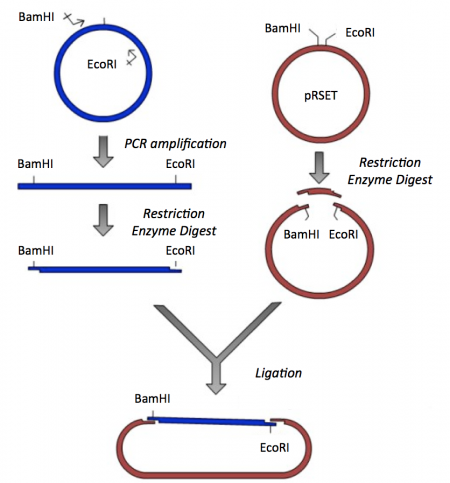
Though the theme of Module 1 is protein engineering, today will focus on a few key techniques used in DNA engineering. Because the sequence of proteins is determined by the sequence of the genes that encode them, learning how to manipulate DNA is an important first step. Today you will complete a cloning reaction to generate a protein expression vector that contains a gene that encodes a calcium-sensing protein. This process is illustrated in the schematic below. Later you will use this construct to engineer a new calcium-sensing protein.
The cloning vector we will use is pRSET. This vector has several features that make it ideal for cloning and protein expression -- both of which are important for this module. The calcium-sensing protein we will study this module is inverse pericam (IPC). We will discuss this protein in much more detail later, for now it is sufficient to know that is used to measure calcium concentrations. To generate your final product you will use three common DNA engineering techniques: PCR amplification, restriction enzyme digestion, and ligation.
PCR amplification
Restriction enzyme digest
Ligation
Next day: Design mutation primers
Previous day: Orientation