Difference between revisions of "20.109(F23):M2D7"
Becky Meyer (Talk | contribs) (→Protocols) |
Noreen Lyell (Talk | contribs) (→Introduction) |
||
| Line 6: | Line 6: | ||
==Introduction== | ==Introduction== | ||
| − | The research goal for this module was to | + | The research goal for this module was to further study small molecules that were previously identified to bind to PfFKBP35. In work completed by the Fall 2022 class, students screened a library of small molecules that were based on the structure of FK506 which is known to bind to PfFKBP35 and the human ortholog, FKBP12. The goal of the initial screen was to examine the binding of each small molecule to PfFKBP35 and to FKBP12 in an effort to identify molecules that could potentially be used to target PfFKBP35 preferentially over FKBP12. From this experiment, the four small molecules used in your research were identified. |
| + | |||
| + | To better characterize the binding parameters of each of the four identified small molecules to PfFKBP35, you examined binding across different concentrations of small molecule. The T<sub>m</sub> for PfFKBP35 across a range of small molecule concentrations can be used to confirm that binding is occurring and can also give some insight into the strength of the binding. If the small molecule binds to FfFKBP35, then a shift in the T<sub>m</sub> will be observed and can be quantified by subtracting the T<sub>m</sub> of PfFKBP35 without small molecule. The strength of binding can qualitatively be addressed by considering the concentrations of small molecule needed to cause a shift in the T<sub>m</sub>. If a shift in T<sub>m</sub> for a small molecule is observed at only high concentrations, then perhaps the strength of binding is not as good as for another small molecule that causes a shift in T<sub>m</sub> at lower concentrations. | ||
| + | |||
| + | |||
| + | |||
| + | |||
One method used to find small molecules that bind a protein of interest is to use a high-throughput screen, such as the SMM. Though we did not use this method as part of this module, we learned about the technology and the data analysis workflow. This technology is an approach used by researchers to screen thousands, to hundreds of thousands, of small molecules in an unbiased manner using commercially available libraries. | One method used to find small molecules that bind a protein of interest is to use a high-throughput screen, such as the SMM. Though we did not use this method as part of this module, we learned about the technology and the data analysis workflow. This technology is an approach used by researchers to screen thousands, to hundreds of thousands, of small molecules in an unbiased manner using commercially available libraries. | ||
Revision as of 19:47, 7 November 2023
Contents
Introduction
The research goal for this module was to further study small molecules that were previously identified to bind to PfFKBP35. In work completed by the Fall 2022 class, students screened a library of small molecules that were based on the structure of FK506 which is known to bind to PfFKBP35 and the human ortholog, FKBP12. The goal of the initial screen was to examine the binding of each small molecule to PfFKBP35 and to FKBP12 in an effort to identify molecules that could potentially be used to target PfFKBP35 preferentially over FKBP12. From this experiment, the four small molecules used in your research were identified.
To better characterize the binding parameters of each of the four identified small molecules to PfFKBP35, you examined binding across different concentrations of small molecule. The Tm for PfFKBP35 across a range of small molecule concentrations can be used to confirm that binding is occurring and can also give some insight into the strength of the binding. If the small molecule binds to FfFKBP35, then a shift in the Tm will be observed and can be quantified by subtracting the Tm of PfFKBP35 without small molecule. The strength of binding can qualitatively be addressed by considering the concentrations of small molecule needed to cause a shift in the Tm. If a shift in Tm for a small molecule is observed at only high concentrations, then perhaps the strength of binding is not as good as for another small molecule that causes a shift in Tm at lower concentrations.
One method used to find small molecules that bind a protein of interest is to use a high-throughput screen, such as the SMM. Though we did not use this method as part of this module, we learned about the technology and the data analysis workflow. This technology is an approach used by researchers to screen thousands, to hundreds of thousands, of small molecules in an unbiased manner using commercially available libraries.
Another method is to design small molecules using a scaffold molecule that is known to bind the protein of interest, such as the approach used in this module. Here we used a set of small molecules that were chemically altered versions of FK506 which is known to bind PfFKBP35. This approach is only useful when a known small molecule binder is known. In addition, using this approach will not be useful in discovering molecules that bind outside of a known binding pocket or active site.
Today we will consider yet another technique used to identify a small molecule that binds to a protein of interest. In this exercise we will think through how a researcher might rationally design a small molecule based on the structure of the protein of interest.
Protocols
Part 1:
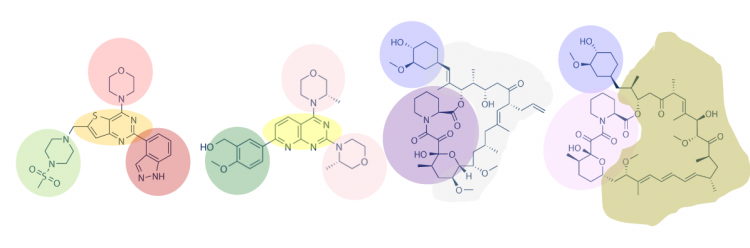
Following an initial screen for putative binders, the small molecules identified as positive hits are then confirmed using functional assays. You will use the PfFKBP35 protein you purified in the next laboratory session to perform a differential scanning fluorimetry (DSF) assay. The DSF assay more directly tests binding between a small molecule and a protein target. In preparation for the DSF experiment, today you will review the structures of the small molecules below and select which you will test using the DSF assay.
One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. Your goal for today is to carefully examine the hits and identify any common features / structures. As in the example image below, it is possible that multiple features will be present within the same small molecule.
In your laboratory notebook, complete the following:
- How many features did you identify that are present in two or more of the small molecules that putatively bind PfFKBP35? Are there more or less than you expected?
- Is there a feature present in all of the identified small molecules? What might this suggest about the binding site(s) and / or binding ability of PfFKBP35?
- Can you assign the identified small molecules to sub-groups based on the common features that are present?
- What might the different features represent? More specifically, consider whether each subgroup has a unique binding site on the target protein or if each subgroup represents different solutions for interacting with the same binding site.
- How might you make modifications to the small molecules / features to probe binding? As a hint, consider how different functional groups could be positioned at a given site without altering qualitative binding in the SMM assay to translate that into some testable ideas (e.g. quantitative binding properties may be occurring that are functionally relevant, but not discernible by SMM assays; or such a site is not critical for binding and may allow for modifications that confer beneficial properties of the compound).
Part 2: other methods for small molecule discovery
A small molecule microarray (SMM) requires the covalent attachment of a library of small molecules to a glass slide. Our library is meant to broadly sample the drug-like chemical space (i.e. all possible chemical structures that have drug-like physical properties) and contains about 50,000 small molecules. Some libraries are much smaller, while many pharmaceutical companies possess high-throughput screening (HTS) collections of millions of compounds. Because this chemical space is very large, it’s difficult to generalize any single chemical reaction for this attachment that can be applied to all small molecules. We take a “one-size-fits-most” approach, where the glass slide is functionalized with a broadly reactive electrophile capable of reacting with nucleophiles present in most drug-like small molecules, such as alcohols or amines. Many small molecules contain multiple nucleophiles suitable for attachment. In this case, our manufacturing will result in a mixture of attachment sites. It’s important to remember that attachment to the glass slide constrains the possible orientation of the protein-small molecule interaction; some orientations are not possible because the glass slide and linker are in the way.
We start with a glass slide with exposed amines across the surface and attach a short PEG (polyethylene glycol) linker. To the end of this PEG linker, an isocyanate group is attached. Isocyanate, or R-N=C=O, is a resonant structure, and a partial positive charge is stabilized on the carbon atom. This carbon atom is electrophilic, and small molecules with nucleophiles will react here. We estimate that about 70% of drug-like small molecules are amenable to this reaction, and our library is filtered to contain only these molecules.
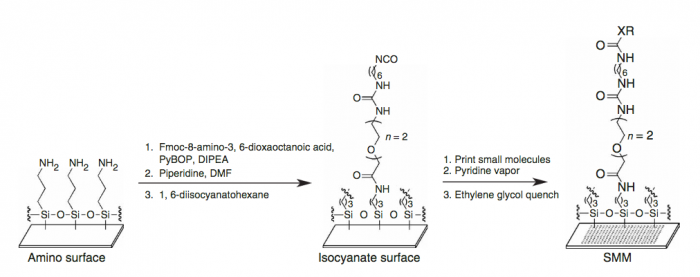
Our compound library is dissolved in DMSO and stored in 384-well plates. To dispense the compounds onto our functionalized glass slide, a robotic arm with a set of 48 metal pins is used to transfer the compounds to the glass slide. Each metal pin has a small slit in the end, and capillary action is used to precisely withdraw and dispense consistent volumes. When the pins touch the glass slide, the compound in DMSO is dispensed into a small circle of approximately 160 micron in diameter. Each pin prints one compound in two different locations on each slide, and then the pins are washed in water and DMSO. This process is repeated for each compound, resulting in our final microarray. The microarray is divided into 48 subarrays, and each subarray corresponds to one pin and contains 256 discrete spots. Within each subarray, we print a set of fluorescent compounds in the shape of an ‘X’ so that we can precisely determine where each spot is printed. After the compounds react, we quench the surface so that no electrophiles remain. This results in our final microarray; a collection of approximately 12,000 discrete spots displaying one compound each.
(Written by Rob Wilson. For more information read Bradner, J. E., McPherson, O. M., and Koehler, A. N. (2006) A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Prot. 1:2344-2352. PMID: 17406478.)
Part 5: Draft discussion section for Research article
As the final section of your Research article, you will write a formal Discussion that summarizes the key findings and states the implications of your research. Use the homework you completed for today to draft the Discussion for your Research article.
Remember that the Results and Discussion information will be separate in this more formal writing assignment. Use the questions below to help you decide which details should be included where in your text.
Next day: Brainstorm ideas for Research Proposal presentation