Difference between revisions of "20.109(F23):M2D6"
Noreen Lyell (Talk | contribs) (→Part 2: Examine binding shifts) |
Noreen Lyell (Talk | contribs) (→Part 2: Examine binding shifts) |
||
| Line 32: | Line 32: | ||
#To determine the T<sub>m</sub> for each concentration tested, visually identify the lowest point on each curve, then use your cursor to identify the values associated with that data point. | #To determine the T<sub>m</sub> for each concentration tested, visually identify the lowest point on each curve, then use your cursor to identify the values associated with that data point. | ||
#*If the T<sub>m</sub> values are unclear from your plot, ask your Instructor how to proceed. | #*If the T<sub>m</sub> values are unclear from your plot, ask your Instructor how to proceed. | ||
| − | #Calculate the & | + | #Calculate the Δ T<sub>m</sub> by quantifying the subtracting the T<sub>m</sub> for the 0 μM sample from the T<sub>m</sub> values determined for each concentration of compound tested. |
#Next, examine the melting curves plotting fluorescence intensity vs. temperature from the "melt" file. Again, validate the results you found by eye to see if the T<sub>m</sub> values correspond to the inflection point of the raw fluorescence melt curves. | #Next, examine the melting curves plotting fluorescence intensity vs. temperature from the "melt" file. Again, validate the results you found by eye to see if the T<sub>m</sub> values correspond to the inflection point of the raw fluorescence melt curves. | ||
#Report the T<sub>m</sub> values for your experiment by entering your results in the table on the Class data tab. | #Report the T<sub>m</sub> values for your experiment by entering your results in the table on the Class data tab. | ||
Revision as of 02:46, 7 November 2023
Contents
Introduction
Today you will analyze the data for the DSF experiment. These results will be the focus of your major writing assignment for this module, the Research article.
As a reminder DSF is a functional assay used to probe the interaction between a single protein of interest and a putative small molecule binder. In this assay, the melting temperature (Tm) of the protein is measured using a fluorescent dye and a change in the melting temperature (ΔTm) when the small molecule is present in the reaction is indicative of binding.
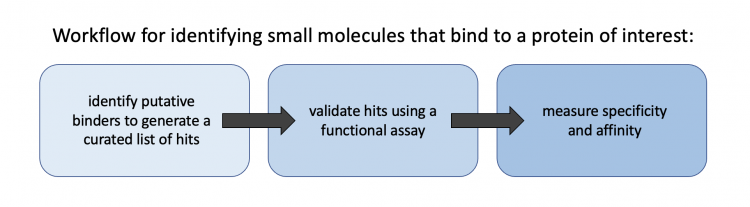
The DSF assay is one method used to validate 'hits' identified using a high-throughput approach, such as the SMM. Though an SMM was not used as part of this module, it was discussed to highlight how a researcher can easily screen through tens-of-thousands of compounds to generate a curated list of small molecules that can be further tested using a functional assay. For this module, we started with a short list of compounds based on known binders of our protein of interest. In either case, the workflow of this type of project is 1) identify putative binders using a screen or other method to generate a curated list of hits, 2) validate hits using a functional assay, 3) measure parameters such as specificity and affinity that exist between the protein of interest and validated hits.
Protocols
Part 1: Participate in Communication Lab workshop
Our communication instructor, Dr. Chiara Ricci-Tam, will join us today for a discussion on preparing formal writing assignments.
Part 2: Examine binding shifts
Your data is provided as two .xlsx files containing raw data from each well of a 96 well plate over the specified range of temperatures (access your files through the link on the Class data tab). The sheet with "Melt" in the file name will contain raw fluorescence intensity data, while the other sheet with "Tm" in its name will have the values for the first derivative of the melt curve. The “X” columns denote the temperature and should the same across the data set. The “B2:Sample ##” columns denote the fluorescence value measured for that well at that temperature. Refer back to your notes to determine what samples were loaded in which wells.
To determine the "melting temperature," or Tm of the protein you will plot the data for each concentration tested and determine the temperature at the inflection point of the melting curve. This inflection point occurs at the maximum value of the first derivative. The BioRad CFX machine we use exports the negative of the first derivative in the Excel file, so we will find the minimum value in the first derivative, then take the corresponding temperature to be the Tm in each condition.
- Each laboratory partner will complete the data analysis for the sample they prepared in the previous session.
- Open the Excel file corresponding to the first derivative data (the "Tm" file).
- First, the data for the triplicate samples that were tested for each concentration must be averaged to attain the value that will be graphed.
- Now, plot the data set by making a scatter plot ("scatter with smooth line and markers") with the temperature on the x-axis and the first derivative values on the y-axis.
- To determine the Tm for each concentration tested, visually identify the lowest point on each curve, then use your cursor to identify the values associated with that data point.
- If the Tm values are unclear from your plot, ask your Instructor how to proceed.
- Calculate the Δ Tm by quantifying the subtracting the Tm for the 0 μM sample from the Tm values determined for each concentration of compound tested.
- Next, examine the melting curves plotting fluorescence intensity vs. temperature from the "melt" file. Again, validate the results you found by eye to see if the Tm values correspond to the inflection point of the raw fluorescence melt curves.
- Report the Tm values for your experiment by entering your results in the table on the Class data tab.
- Last, compare your results to the positive and negative control compounds. You can find the graphed results for rapamycin (positive control) and pyrimethamine (negative control) in the folder named 'RA data'.
In your laboratory notebook, complete the following:
- Record the Tm values.
- Attach the graphs generated as part of the analysis.
Part 3: Organize figures and outline results text for your Research article
The goal for today is to focus on how you will communicate the results you are gathering and analyzing in the Research article.
Currently, you have partial drafts and outlines for each of the sections (with Instructor feedback!) that will be included in the Research article. Today you will organize and write a detailed outline for the data that you collected for this module. Use the skills you learned when you completed the figure homework and apply them to the remaining figures for your Research article.
To get started on this process, complete the following:
- Make a list of all of the schematics / data figures / tables that will be included.
- Organize the figures such that the data tell a coherent story that answers your research question.
- Complete the following steps for each figure:
- Write a conclusive figure title that relays the main take-home message for the data shown.
- Write a results subsection title that mimics the figure title.
- Outline the text that will be included in the results subsection using the prompts included in the Results section of Research article page.
In your laboratory notebook, complete the following:
- Include the list of schematics / data figures / tables.
- For each schematic / data figure / table, provide a brief summary of the information that will included in the text of the results section.
Next day: Organize results and incorporate class data