20.109(F22):M2D2
Introduction
To induce production of PfFKBP35 protein from the expression plasmid that was 'cloned' in the previous laboratory session, rhamnose was used to induce expression in KRX E. coli bacterial cells. The use of rhamnose to induce protein expression is based on the native rha operon used for rhamnose metabolism in bacterial cells.
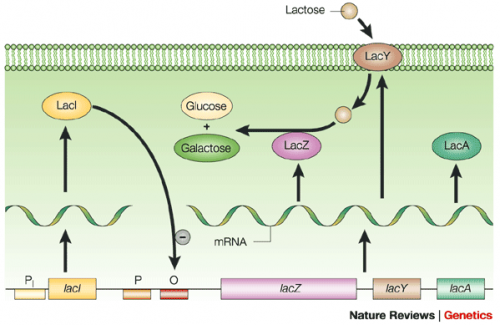
The lac operon is composed of four genes: lacI, lacZ, lacY, and lacA. When lactose is absent, LacI (the protein encoded by lacI) binds to the operator sequence (O) upstream of lacZYA. In the presence of lactose, LacI and lactose form a complex which relieves repression of lacZYA transcription. LacZ is a β-galactosidase that cleaves lactose resulting in glucose and galactose. LacY, a β-galactoside permease, facilitates the transport of lactose across the cell membrane, and LacA, a β-galactoside transacetylase, transfers an acetyl group from acetyl-CoA to β-galactosides.The native rha operon is a powerful tool in engineering protein expression systems because it enables researchers to control gene expression using inducer molecules. The rhaSR genes are only expressed when lactose is present. If a gene of interest is cloned downstream of the operator sequence, the expression of this gene can be controlled by RhaS repression and rhamnose derepression. To further control the system for protein expression, the genes responsible for metabolism of rhamnose are not present in KRX E. coli cells and therefore the molecule is not consumed for growth.
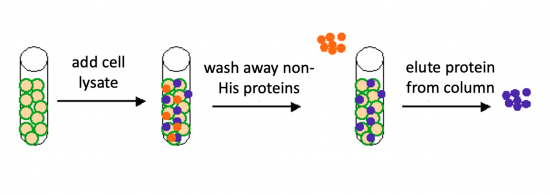
Today you will isolate the expressed PfFKBP35 protein from the bacterial cells. Remember that the PfFKBP35 gene sequence was synthesized such that a 6xHis-tag was added to the DNA sequence. The resultant protein is therefore His-tagged. Histidine has several interesting properties, notably its near-neutral pKa, and His-rich peptides are promiscuous binders, particularly to metals. (For example, histidine side chains help coordinate iron molecules in hemoglobin.)
To purify the PfFKBP35 proteins present in the bacterial cell, you will use a nickel-agarose resin. The 6xHis-tag on the PfFKBP35 protein will bind to the nickel-coated resin, while the other cellular protein will pass through the resin. Remember, the KRX cells are not only producing the PfFKBP35 protein, but also the proteins needed for cellular function and survival. Imidazole is a compound that is also able to bind to nickel and washing the resin with a low concentration solution promotes competition for binding between the imidazole and bound proteins for the nickel-coated resin. Proteins that are non-specifically bound will have a lower affinity for the nickel than imidazole and be washed from the column, whereas the 6xHis-tagged PfFKBP35 will remain adhered to the nickel-agarose resin. To elute the PfFKBP35 protein from the nickel-coated resin, a high concentration of imidazole is used to out-compete the 6xHis-tag for binding.
Protocols
Part 1: Induce expression of PfFKBP35
For timing reasons, the induction steps were completed prior to class. So you understand how the cell pellets you will use for the protein purification, the steps are shown in the video and protocol below.
To ensure the steps included below are clear, please watch the video tutorial linked here: [Bacterial Induction]. The steps are detailed below so you can follow along!
- Inoculated 5 mL of TB media containing 50 μg/mL kanamycin with a colony of Nico(DE3) cells transformed with pET-28a_FKBP35.
- Incubated the culture overnight at 37 °C with shaking at 220 rpm.
- Dilute the overnight culture 1:100 in 1 L of fresh TB media containing 50 μg/mL kanamycin.
- Incubate at 37 °C until the OD600 = 0.6 with shaking at 220 rpm.
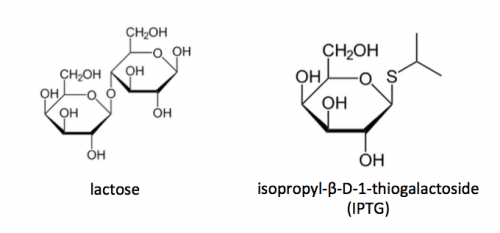
- To induce PfFKBP35 protein expression, add IPTG to a final concentration of 1 mM.
- Incubate at overnight at room temperature with shaking at 100 rpm.
- To harvest the cells, centrifuge the culture at 4000 g for 15 min at 4 °C.
- Cell pellets were flash frozen in liquid nitrogen, then stored at -80 °C until used for purification.
In your laboratory notebook, complete the following:
- Calculate the volume of kanamycin stock that was added to the TB broth in Step #1. In Step #3.
- Concentration of kanamycin stock = 50 mg/mL.
- Calculate the volume of IPTG stock that was added to the TB broth in Step #5.
- Concentration of IPTG stock = 100 mM.
Part 2: Purify PF3D7_20109-F21 protein
To ensure the steps required for purifying the PfFKBP35 protein are clear, the Instructor will provide a live demonstration of this process.
Lyse Nico(DE3) cells expressing pET-28a_FKBP35
- Retrieve 4 pellets of the Nico(DE3) pET-28a_FKBP35 cell pellet from the -80 °C freezer and leave it on your bench to thaw.
- To lyse the cells, add B-Per bacterial extraction reagent at 2 mL / cell pellet.
- Resuspend the cell pellets in the lysis buffer then combine such that one tube contains the contents of 4 cell pellets.
- To the pooled tube add:
- lysozyme at 2 μL / mL of B-Per bacterial extraction reagent
- 1 protease inhibitor tablet
- Solubilize the cell pellet in lysis buffer and vortex to mix.
- Incubate cell pellet in lysis buffer at room temperature for 15 min on nutator.
- To pellet the cell debris, centrifuge the lysate at 3,000 rcf for 45 min at 4 °C.
- Complete Part 3: Electrophorese confirmation digests during the centrifugation.
Prepare Strep-Tactin affinity column
- Obtain a 400 μL aliquot of 50% Strep-Tactin resin.
- Transfer the resin to an column.
- Immediately prior to adding cell lysate, wash the resin.
- To wash the resin, pass 200 μL of 1X PBS through the column
- Be sure a container is placed under the column to collect the waste!
- When the PBS has flowed through, cap the bottom and the top of the column until you are ready to add the cell lysate.
Purify PF3D7_1351100 from cell lysate
- Transfer the supernatent from the centrifuged cell lysate to a fresh conical tube.
- Label the microcentrifuge tube containing the cell pellet as "pellet" and give it to the Instructor! This pellet will be used later when protein expression and purity are examined.
- Aliquot 30 μL of the supernatent (from Step #1) to a fresh microcentrifuge tube.
- Label the microcentrifuge tube containing the aliquot as "lysate" and give it to the Instructor! This aliquot will be used later when protein expression and purity are examined.
- Pipet 3 mL of supernatent into the prepared Strep-Tactin affinity column.
- Be sure that the bottom of the column is capped!
- Pipette the resin / cell supernatent to mix and incubate at room temperature for 5 min.
- Hold a microcentrifuge tube under the column, then remove the bottom cap from the column and collect the liquid that leaves the column.
- Label the microcentrifuge tube as "flowthrough" and give it to the Instructor! This aliquot will be used later when protein expression and purity are examined.
- If you have more than 3 mL supernatent, repeat steps #3 - 4. Flowthrough only needs to be collected from the first round.
- To wash the Strep-Tactin affinity column, add 10 mL of wash buffer (1x PBS).
- Hold a microcentrifuge tube under the column, then remove the bottom cap from the column and collect ~250 μL of the liquid that leaves the column.
- Label the microcentrifuge tube as "wash" and give it to the Instructor! This aliquot will be used later when protein expression and purity are examined.
- To elute the PF3D7_20109-F21 protein from the affinity column, add 1 mL of elution buffer and incubate at room temperature for 5 min.
- Hold a microcentrifuge tube under the column, then remove the bottom cap from the column and collect the entire 1 mL of the liquid that leaves the column.
- Label the microcentrifuge tube as "elution" and give it to the Instructor! This aliquot will be used later when protein expression and purity are examined.
- Lastly, resuspend the slurry from the Strep-Tactin affinity column in 250 μL 1X PBS and transfer to a fresh microcentrifuge tube.
- Label the microcentrifuge tube as "slurry" and give it to the Instructor! This aliquot will be used later when protein expression and purity are examined.
In your laboratory notebook, complete the following:
- At several steps in the protein purification procedure, samples are collected that will be used later when protein expression and purity are examined. Consider why each of the samples listed below are saved as controls to measure the success of the purification.
- The pellet from Step #1.
- The lysate from Step #2.
- The flowthrough from Step #6.
- The wash from Step #7.
- The slurry from Step #9.
- What is occurring during the incubation in Step #4?
Part 3: Electrophorese confirmation digests
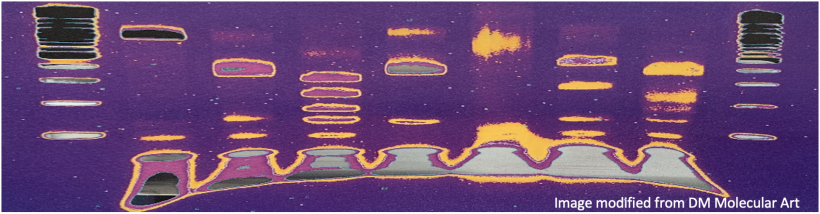
Electrophoresis is a technique that separates large molecules by size using an applied electrical field and a sieving matrix. DNA, RNA and proteins are the molecules most often studied with this technique; agarose and acrylamide gels are the two most common sieves. The molecules to be separated enter the matrix through a well at one end and are pulled through the matrix when a current is applied across it. The larger molecules get entwined in the matrix and are stalled; the smaller molecules wind through the matrix more easily and travel farther away from the well. The distance a DNA fragment travels is inversely proportional to the log of its length. Over time fragments of similar length accumulate into “bands” in the gel. Higher concentrations of agarose can be used to resolve smaller DNA fragments.
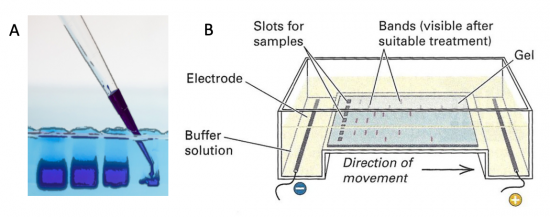
DNA and RNA are negatively charged molecules due to their phosphate backbone, and they naturally travel toward the positive electrode at the far end of the gel. Today you will separate DNA fragments using an agarose matrix. Agarose is a polymer that comes from seaweed. To prepare these gels, agarose and 1X TAE buffer (Tris base, acetic acid, and EDTA) are microwaved until the agarose is melted and fully dissolved. The molten agar is then poured into a horizontal casting tray, and a comb is added. Once the agar has solidified, the comb is removed, leaving wells into which the DNA samples can be loaded.
For the digests that were prepared in the previous laboratory session, a 1% agarose gel with SYBR Safe DNA stain was used to separate the DNA fragments in the four digest reactions. In addition, a well was loaded with a molecular weight marker (also called a DNA ladder) to determine the size of the fragments.
To ensure the steps included below are clear, please watch the video tutorial linked here: [DNA gel electrophoresis].
- Add 5 μL of 6x loading dye to the digests.
- Loading dye contains bromophenol blue as a tracking dye, which enables you to follow the progress of the electrophoresis.
- Glycerol is also included to weight the samples such that the liquid sinks into well.
- Flick the eppendorf tubes to mix the contents, then quick spin them in the microfuge to bring the contents of the tubes to the bottom.
- Load 25 μL of each digest into the gel, as well as 5 μL of 1kb DNA ladder.
- Be sure to record the order in which you load your samples!
- To load your samples, draw the volume listed above into the tip of your P200 or P20. Lower the tip below the surface of the buffer and directly over the well. Avoid lowering the tip too far into the well itself so as to not puncture the well. Expel your sample slowly into the well. Do not release the pipet plunger until after you have removed the tip from the gel box (or you'll draw your sample back into the tip!).
- Once all the samples have been loaded, attach the gel box to the power supply and electrophorese the gel at 125 V for 45 minutes.
- Lastly, visualize the DNA fragments in the agarose gel using the gel documentation system.
Reagents list
- Terrific broth (TB) (from RPI)
- kanamycin (from Sigma)
- isopropyl β-d-1-thiogalactopyranoside (IPTG) (from Sigma)
- 2x B-Per bacterial protein extraction reagent (from ThermoFisher)
- lysozyme (from Sigma)
- protease inhibitor tablet (from Sigma)
- phosphate saline buffer (PBS) (from VWR)
- Strep-Tactin agarose (from IBA Life Sciences)
- Wash buffer: 1X PBS
- Elution buffer: 1X PBS containing 2.5 mM biotin
Next day: Assess purity and concentration of purified protein