20.109(F20):M2D1
Introduction
Though the theme of Module 1 is focused on screening for small molecules that bind PROTEIN X, today will focus on a few key techniques used in DNA engineering. Because the sequence of proteins is determined by the sequence of the genes that encode them, learning how to manipulate DNA is an important first step. Today you will complete the cloning steps used to incorporate the gene that encodes the PF3D7_1351100 protein into an expression vector. The expression vector contains the genetic elements needed to express and purify of a protein of interest.
[[|thumb|center|450px|Schematic of pET-28b(+)_PF3D7_1351100 cloning. First, the TDP43 insert is PCR amplified to generate multiple copies of the fragment that are flanked by restriction enzymes sites. Next, this fragment and the vector are digested to create compatible ends. Last, the compatible ends of the digested insert and vector are 'glued together' in a ligation reaction.]]
The vector has several features that make it ideal for cloning and plasmid replication -- both of which are important for this module. To generate your final product you will use three common DNA engineering techniques: PCR amplification, restriction enzyme digestion, and ligation.
Restriction enzyme digest
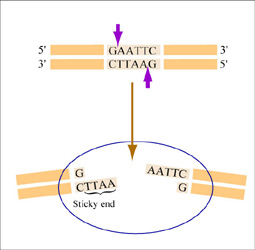
Restriction endonucleases, also called restriction enzymes, 'cut' or 'digest' DNA at specific sequences of bases. The restriction enzymes are named according to the prokaryotic organism from which they were isolated. For example, the restriction endonuclease EcoRI (pronounced “echo-are-one”) was originally isolated from E. coli giving it the “Eco” part of the name. “RI” indicates the particular version on the E. coli strain (RY13) and the fact that it was the first restriction enzyme isolated from this strain.
The sequence of DNA that is bound and cleaved by an endonuclease is called the recognition sequence or restriction site. These sequences are usually four or six base pairs long and palindromic, that is, they read the same 5’ to 3’ on the top and bottom strand of DNA. For example, the recognition sequence for EcoRI is below (see also figure at right). EcoRI cleaves the phosphate backbone of DNA between the G and A of the recognition sequence, which generates overhangs or 'sticky ends' of double-stranded DNA.
5’ GAATTC 3’
3’ CTTAAG 5’
Unlike EcoRI, some other restriction enzymes cut precisely in the middle of the palindromic DNA sequence, thus leaving no overhangs after digestion. The single-stranded overhangs resulting from DNA digestion by enzymes such as EcoRI are called sticky ends, while double-stranded ends resulting from digestion by enzymes such as HaeIII are called blunt ends. HaeIII recognizes
5’ GGCC 3’
3’ CCGG 5’
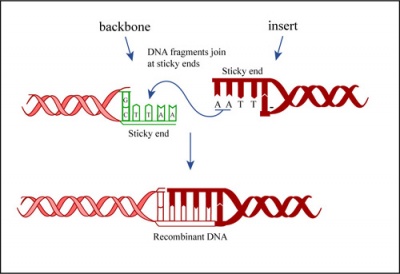
Ligation
In a ligation reaction, DNA ends are covalently attached to one another via the ligase enzyme. The efficiency of the reaction is related to type of DNA ends: compatible sticky ends will ligate more efficiently than blunt ends, and non-compatible sticky ends will not be ligated due to the lack of hydrogen bonding between the basepairs. To initiate the ligation reaction, hydrogen bonds are formed between the compatible overhangs of DNA fragments. The ligase enzyme then forms a covalent phosphodiester bond between the 3' hydroxyl end of the 'acceptor' nucleotide and the 5' phosphodiester end of the 'donor' nucleotide.
The first step in this process is the addition of AMP (adenylation) to a lysine residue within the active site of DNA ligase, which releases a pyrophosphate. Next, the AMP is transferred to the 5' phosphate of the donor nucleotide resulting in the formation of a pyrophosphate bond. Lastly, a phosphodiester bond is formed between the 5' phosphate of the donor nucleotide and the 3' hydroxyl of the 3' acceptor nucleotide.