20.109(F17):Test role of biochemical factors in genomic stability (Day4)
Contents
Introduction
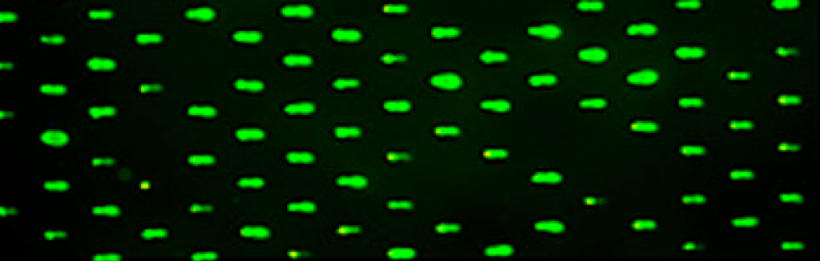
In the initial experiment for this module, your goal was to optimize the CometChip assay. Now that you have data concerning how to best load cells into the CometChip, you will use this technique to study the Base Excision Repair (BER) pathway. BER corrects DNA damage, specifically base lesions, that result from oxidation, alkylation, deamination, and depuriniation/depryrimidination. If base lesions are not repaired, non-canonical base pairing can occur, which may result in the incorporation of an incorrect base during replication. To prevent mutations and maintain integrity of the genome, the BER pathway evolved as a highly conserved repair mechanism in both E. coli and mammals.
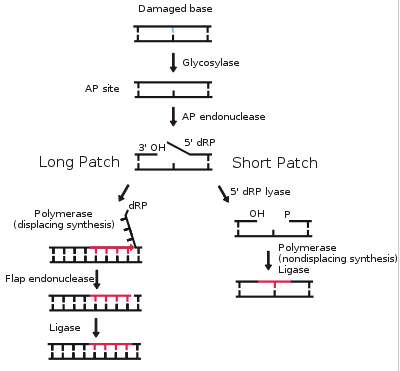
The core BER pathway includes only four proteins that function to remove the damaged base and replace it with the correct base. First, a DNA glycosylase recognizes that a damaged base is present in the DNA and cleaves an N-glycosidic bond, which creates an apurinic or apyrimidinic site (referred to as an AP site in both cases). Different DNA glycosylases recognize different types of base lesions. Second, the DNA backbone is cleaved to create a single-strand DNA nick by either a DNA AP endonuclease or a DNA AP lyase. Next, a DNA polymerase incorporates the correct base using the sister strand as a template. Last, a DNA ligase completes the repair by sealing the single-strand nick, which restores integrity to the helix. For a more detailed description of the BER pathway, read this review by Robertson et al.The BER pathway repairs base lesions using either a short-patch or long-patch mechanism. In the short-patch BER pathway, a single nucleotide is removed in the repair process. In contrast, in the long-patch BER pathway at least two nucleotides are removed. Though both mechanisms of repair are utilized, the conditions that drive each are poorly understood.
To study the BER pathway, as a class you will use two chemical treatments to generate base lesions. The treatments will be applied to two CometChips as a second variable will be introduced during the next laboratory session.
mutant cell lines!!!
Protocols
Part 1: Load CometChips to test biochemical factors
- You will load the CometChips you prepared during the previous laboratory session using the procedure from M1D2.
- For the cell number, you will use the values decided on during the group discussion on M1D3.
- The cell suspension prepared by the teaching faculty is at a density of XX cells / mL.
After you load the cells into the CometChip, you will induce DNA damage using either H2O2 or MMS. Each team will select one of the chemical treatments on the sign-up sheet located at the front laboratory bench.
Follow the appropriate protocol for damage induction based on the chemical you were assigned to test.
Include information re: different cell types
Part 2: Induce DNA damage using H2O2 or MMS treatments
Part 3: Complete cell lysis
Reagents list
Next day: Complete biochemical experiment